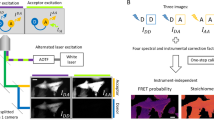
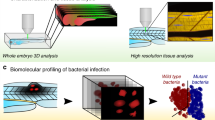
Abstract
Fluorescence resonance energy transfer (FRET)-based molecular biosensors serve as important tools for studying protein activity in live cells and have been widely used for this purpose over the past decade. However, FRET biosensors are rarely used in the context of the live organism because of the inherent high cellular complexity and imaging challenges associated with the three-dimensional environment. Here we provide a protocol for using single-chain intramolecular FRET-based biosensors in early development. We provide a general protocol for FRET ratio imaging in embryos, including the data-acquisition conditions and the algorithm for ratio image generation. We then use the pRaichu RacFRET biosensor to exemplify the adaptation and optimization of a particular biosensor for use in live zebrafish embryos. Once an optimized biosensor is available, the complete procedure, including introduction of the probes into embryos, imaging and data analysis, requires 2–3 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
References
Palmer, A.E., Qin, Y., Park, J.G. & McCombs, J.E. Design and application of genetically encoded biosensors. Trends Biotechnol. 29, 144–152 (2011).
Merlot, S. & Firtel, R.A. Leading the way: directional sensing through phosphatidylinositol 3-kinase and other signaling pathways. J. Cell Sci. 116, 3471–3478 (2003).
Devreotes, P. & Janetopoulos, C. Eukaryotic chemotaxis: distinctions between directional sensing and polarization. J. Biol. Chem. 278, 20445–20448 (2003).
Tsien, R.Y. & Miyawaki, A. Seeing the machinery of live cells. Science (New York, NY) 280, 1954–1955 (1998).
Jares-Erijman, E.A. & Jovin, T.M. FRET imaging. Nat. Biotechnol. 21, 1387–1395 (2003).
Kiyokawa, E., Hara, S., Nakamura, T. & Matsuda, M. Fluorescence (Forster) resonance energy transfer imaging of oncogene activity in living cells. Cancer Sci. 97, 8–15 (2006).
VanEngelenburg, S.B. & Palmer, A.E. Fluorescent biosensors of protein function. Curr. Opin. Chem. Biol. 12, 60–65 (2008).
Kraynov, V.S. et al. Localized Rac activation dynamics visualized in living cells. Science (New York, NY) 290, 333–337 (2000).
Pertz, O., Hodgson, L., Klemke, R.L. & Hahn, K.M. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature 440, 1069–1072 (2006).
Gaits, F. & Hahn, K. Shedding light on cell signaling: interpretation of FRET biosensors. Sci. STKE 2003, PE3 (2003).
Aoki, K. & Matsuda, M. Visualization of small GTPase activity with fluorescence resonance energy transfer-based biosensors. Nat. Protoc. 4, 1623–1631 (2009).
Esposito, A., Gralle, M., Dani, M.A., Lange, D. & Wouters, F.S. pHlameleons: a family of FRET-based protein sensors for quantitative pH imaging. Biochemistry 47, 13115–13126 (2008).
Miyawaki, A. et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388, 882–887 (1997).
Palmer, A.E. & Tsien, R.Y. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat. Protoc. 1, 1057–1065 (2006).
Itoh, R.E. et al. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol. Cell Biol. 22, 6582–6591 (2002).
Wang, Y. et al. Visualizing the mechanical activation of Src. Nature 434, 1040–1045 (2005).
Fosbrink, M., Aye-Han, N.N., Cheong, R., Levchenko, A. & Zhang, J. Visualization of JNK activity dynamics with a genetically encoded fluorescent biosensor. Proc. Natl. Acad. Sci. USA 107, 5459–5464 (2010).
Sato, M., Kawai, Y. & Umezawa, Y. Genetically encoded fluorescent indicators to visualize protein phosphorylation by extracellular signal-regulated kinase in single living cells. Anal. Chem. 79, 2570–2575 (2007).
Kiyokawa, E., Aoki, K., Nakamura, T. & Matsuda, M. Spatiotemporal regulation of small GTPases as revealed by probes based on the principle of Forster Resonance Energy Transfer (FRET): implications for signaling and pharmacology. Ann. Rev. Pharmacol. Toxicol. 51, 337–358 (2011).
Kardash, E. et al. A role for Rho GTPases and cell-cell adhesion in single-cell motility in vivo. Nat. Cell Biol. 12, 47–53; sup pp 1–11 (2010).
Miyagi, C. et al. STAT3 noncell-autonomously controls planar cell polarity during zebrafish convergence and extension. J. Cell Biol. 166, 975–981 (2004).
Stockholm, D. et al. Imaging calpain protease activity by multiphoton FRET in living mice. J. Mol. Biol. 346, 215–222 (2005).
Blaser, H. et al. Migration of zebrafish primordial germ cells: a role for Myosin contraction and cytoplasmic flow. Dev. Cell 11, 613–627 (2006).
Kamiyama, D. & Chiba, A. Endogenous activation patterns of Cdc42 GTPase within Drosophila embryos. Science (New York, NY) 324, 1338–1340 (2009).
Wang, X., He, L., Wu, Y.I., Hahn, K.M. & Montell, D.J. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell Biol. 12, 591–597 (2010).
Horikawa, K. et al. Spontaneous network activity visualized by ultrasensitive Ca(2+) indicators, yellow Cameleon-Nano. Nat. Methods 7, 729–732 (2010).
Xia, Z. & Liu, Y. Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys. J. 81, 2395–2402 (2001).
Nakamura, T., Aoki, K. & Matsuda, M. Monitoring spatio-temporal regulation of Ras and Rho GTPase with GFP-based FRET probes. Methods 37, 146–153 (2005).
Köprunner, M., Thisse, C., Thisse, B. & Raz, E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 15, 2877–2885 (2001).
Goedhart, J. & Kremers, G.-J. in Laboratory Techniques in Biochemistry and Molecular Biology, FRET and FLIM Techniques, Vol. 33 (ed. Gadella, T.W.) Ch. 5, 171–223 (Elsevier, 2009).
Nguyen, A.W. & Daugherty, P.S. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 23, 355–360 (2005).
Shaner, N.C., Steinbach, P.A. & Tsien, R.Y. A guide to choosing fluorescent proteins. Nat. Methods 2, 905–909 (2005).
Ouyang, M., Sun, J., Chien, S. & Wang, Y. Determination of hierarchical relationship of Src and Rac at subcellular locations with FRET biosensors. Proc. Natl. Acad. Sci. USA 105, 14353–14358 (2008).
Diekmann, D. et al. Bcr encodes a GTPase-activating protein for p21rac. Nature 351, 400–402 (1991).
Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of zebrafish (Danio rerio), 4th edn. (University of Oregon Press, 2000).
Rosen, J.N., Sweeney, M.F. & Mably, J.D. Microinjection of zebrafish embryos to analyze gene function. J. Vis. Exp. published online, doi:10.3791/1115 (9 March 2009).
Griesbeck, O., Baird, G.S., Campbell, R.E., Zacharias, D.A. & Tsien, R.Y. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J. Biol. Chem. 276, 29188–29194 (2001).
Acknowledgements
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) and the Max Planck Society. We thank M. Reichman-Fried for critical reading of the manuscript and E.-M. Messerschmidt for technical assistance. We also thank M. Matsuda and P. Daugherty for sharing the constructs.
Author information
Authors and Affiliations
Contributions
E.K. designed and performed the experiments, analyzed the data and wrote the paper. J.B. designed and performed the experiments, analyzed the data and wrote the paper. E.R. supervised the project, designed the experiments and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figure 1
Raw data captured in the YFP channel, of a cell expressing SECFP-YPet construct with the histogram showing the pixel intensity distribution. The maximum intensity value detected by the camera is 4095. To generate the histogram, the 16 bit image is first converted to 32 bit using the command: Image > Image Type > 32 bit from the ImageJ menu. To obtain the histogram, the command: Analyse > Histogram was used and the desired range was defined in the Histogram dialog box. (JPG 460 kb)
Supplementary Data 1
rawCFP image. The CFP emission channel of the somatic cell expressing RacFRET-noCT construct. (It is recommended that this file is viewed using ImageJ, a public domain Java image processing program available at http://rsbweb.nih.gov/ij/) (TIFF 51 kb)
Supplementary Data 2
rawYFP image. The YFP emission channel of the somatic cell expressing RacFRET-noCT construct. (It is recommended that this file is viewed using ImageJ, a public domain Java image processing program available at http://rsbweb.nih.gov/ij/) (TIFF 51 kb)
Supplementary Table 1
List of example DNA constructs used in this protocol (DOC 964 kb)
Rights and permissions
About this article
Cite this article
Kardash, E., Bandemer, J. & Raz, E. Imaging protein activity in live embryos using fluorescence resonance energy transfer biosensors. Nat Protoc 6, 1835–1846 (2011). https://doi.org/10.1038/nprot.2011.395
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.395
This article is cited by
-
RETRACTED ARTICLE: A fast, high-affinity fluorescent serotonin biosensor engineered from a tick lipocalin
Nature Methods (2021)
-
Unjamming overcomes kinetic and proliferation arrest in terminally differentiated cells and promotes collective motility of carcinoma
Nature Materials (2019)
-
Aurora-A mediated phosphorylation of LDHB promotes glycolysis and tumor progression by relieving the substrate-inhibition effect
Nature Communications (2019)
-
Fluorescence Polarization of Methylene Blue as a Quantitative Marker of Breast Cancer at the Cellular Level
Scientific Reports (2019)
-
Extra-skeletal manifestations in mice affected by Clcn7-dependent autosomal dominant osteopetrosis type 2 clinical and therapeutic implications
Bone Research (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.