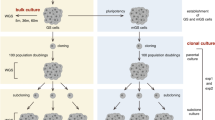
Abstract
We describe an experimental approach for generating mutant alleles in rat spermatogonial stem cells (SSCs) using Sleeping Beauty (SB) transposon–mediated insertional mutagenesis. The protocol is based on mobilization of mutagenic gene-trap transposons from transfected plasmid vectors into the genomes of cultured stem cells. Cells with transposon insertions in expressed genes are selected on the basis of activation of an antibiotic-resistance gene encoded by the transposon. These gene-trap clones are transplanted into the testes of recipient males (either as monoclonal or polyclonal libraries); crossing of these founders with wild-type females allows the insertions to be passed to F1 progeny. This simple, economic and user-friendly methodological pipeline enables screens for functional gene annotation in the rat, with applicability in other vertebrate models where germ line–competent stem cells have been established. The complete protocol from transfection of SSCs to the genotyping of heterozygous F1 offspring that harbor genomic SB gene-trap insertions takes 5–6 months.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Jacob, H.J., Lazar, J., Dwinell, M.R., Moreno, C. & Geurts, A.M. Gene targeting in the rat: advances and opportunities. Trends Genet. 26, 510–518 (2010).
Izsvák, Z. et al. Generating knockout rats by transposon mutagenesis in spermatogonial stem cells. Nat. Methods 7, 443–445 (2010).
Ivics, Z. et al. Transposon-mediated genome manipulation in vertebrates. Nat. Methods 6, 415–422 (2009).
Ivics, Z. & Izsvak, Z. The expanding universe of transposon technologies for gene and cell engineering. Mob. DNA 1, 25 (2010).
Miskey, C., Izsvak, Z., Plasterk, R.H. & Ivics, Z. The Frog Prince: a reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res. 31, 6873–6881 (2003).
Zagoraiou, L. et al. In vivo transposition of Minos, a Drosophila mobile element, in mammalian tissues. Proc. Natl. Acad. Sci. USA 98, 11474–11478 (2001).
de Wit, T. et al. Tagged mutagenesis by efficient Minos-based germ line transposition. Mol. Cell Biol. 30, 68–77 (2010).
Ding, S. et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483 (2005).
Wilson, M.H., Coates, C.J. & George, A.L. Jr. PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 15, 139–145 (2007).
Kawakami, K., Shima, A. & Kawakami, N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc. Natl. Acad. Sci. USA 97, 11403–11408 (2000).
Dupuy, A.J., Fritz, S. & Largaespada, D.A. Transposition and gene disruption in the male germline of the mouse. Genesis 30, 82–88 (2001).
Horie, K. et al. Efficient chromosomal transposition of a Tc1/mariner-like transposon Sleeping Beauty in mice. Proc. Natl. Acad. Sci. USA 98, 9191–9196 (2001).
Keng, V.W. et al. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat. Methods 2, 763–769 (2005).
Suzuki, N. & Withers, H.R. Exponential decrease during aging and random lifetime of mouse spermatogonial stem cells. Science 202, 1214–1215 (1978).
Hamra, F.K. et al. Self renewal, expansion, and transfection of rat spermatogonial stem cells in culture. Proc. Natl. Acad. Sci. USA 102, 17430–17435 (2005).
Ryu, B.Y., Kubota, H., Avarbock, M.R. & Brinster, R.L. Conservation of spermatogonial stem cell self-renewal signaling between mouse and rat. Proc. Natl. Acad. Sci. USA 102, 14302–14307 (2005).
Buehr, M. et al. Capture of authentic embryonic stem cells from rat blastocysts. Cell 135, 1287–1298 (2008).
Li, P. et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell 135, 1299–1310 (2008).
Hamra, F.K. et al. Production of transgenic rats by lentiviral transduction of male germ-line stem cells. Proc. Natl. Acad. Sci. USA 99, 14931–14936 (2002).
Kanatsu-Shinohara, M. et al. Production of transgenic rats via lentiviral transduction and xenogeneic transplantation of spermatogonial stem cells. Biol. Reprod. 79, 1121–1128 (2008).
Ryu, B.Y. et al. Efficient generation of transgenic rats through the male germline using lentiviral transduction and transplantation of spermatogonial stem cells. J. Androl. 28, 353–360 (2007).
Kanatsu-Shinohara, M. et al. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol. Reprod. 69, 612–616 (2003).
Kanatsu-Shinohara, M., Toyokuni, S. & Shinohara, T. Genetic selection of mouse male germline stem cells in vitro: offspring from single stem cells. Biol. Reprod. 72, 236–240 (2005).
Kanatsu-Shinohara, M. et al. Production of knockout mice by random or targeted mutagenesis in spermatogonial stem cells. Proc. Natl. Acad. Sci. USA 103, 8018–8023 (2006).
Brinster, R.L. & Avarbock, M.R. Germline transmission of donor haplotype following spermatogonial transplantation. Proc. Natl. Acad. Sci. USA 91, 11303–11307 (1994).
Hamra, F.K. et al. Defining the spermatogonial stem cell. Dev. Biol. 269, 393–410 (2004).
Russell, W.L. et al. Specific-locus test shows ethylnitrosourea to be the most potent mutagen in the mouse. Proc. Natl. Acad. Sci. USA 76, 5818–5819 (1979).
Driever, W. et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123, 37–46 (1996).
Haffter, P. et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123, 1–36 (1996).
Kile, B.T. et al. Functional genetic analysis of mouse chromosome 11. Nature 425, 81–86 (2003).
Smits, B.M. & Cuppen, E. Rat genetics: the next episode. Trends Genet. 22, 232–240 (2006).
Bushman, F.D. Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell 115, 135–138 (2003).
Jahner, D. et al. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature 298, 623–628 (1982).
Jahner, D. & Jaenisch, R. Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivity. Nature 315, 594–597 (1985).
Carlson, C.M. et al. Transposon mutagenesis of the mouse germline. Genetics 165, 243–256 (2003).
Fischer, S.E., Wienholds, E. & Plasterk, R.H. Regulated transposition of a fish transposon in the mouse germ line. Proc. Natl. Acad. Sci. USA 98, 6759–6764 (2001).
Horie, K. et al. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol. Cell Biol. 23, 9189–9207 (2003).
Yae, K. et al. Sleeping beauty transposon-based phenotypic analysis of mice: lack of Arpc3 results in defective trophoblast outgrowth. Mol. Cell Biol. 26, 6185–6196 (2006).
Geurts, A.M. et al. Gene mutations and genomic rearrangements in the mouse as a result of transposon mobilization from chromosomal concatemers. PLoS Genet. 2, e156 (2006).
Kitada, K. et al. Transposon-tagged mutagenesis in the rat. Nat. Methods 4, 131–133 (2007).
Lu, B. et al. Generation of rat mutants using a coat color-tagged Sleeping Beauty transposon system. Mamm. Genome 18, 338–346 (2007).
Wang, W., Bradley, A. & Huang, Y. A piggyBac transposon-based genome-wide library of insertionally mutated Blm-deficient murine ES cells. Genome Res. 19, 667–673 (2009).
Tong, C., Li, P., Wu, N.L., Yan, Y. & Ying, Q.L. Production of p53 gene knockout rats by homologous recombination in embryonic stem cells. Nature 467, 211–213 (2010).
Urnov, F.D., Rebar, E.J., Holmes, M.C., Zhang, H.S. & Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646 (2010).
Meng, X., Noyes, M.B., Zhu, L.J., Lawson, N.D. & Wolfe, S.A. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 26, 695–701 (2008).
Geurts, A.M. et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science 325, 433 (2009).
Hansen, G.M. et al. Large-scale gene trapping in C57BL/6N mouse embryonic stem cells. Genome Res. 18, 1670–1679 (2008).
Stanford, W.L., Cohn, J.B. & Cordes, S.P. Gene-trap mutagenesis: past, present and beyond. Nat. Rev. Genet. 2, 756–768 (2001).
Friedrich, G. & Soriano, P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 5, 1513–1523 (1991).
Zambrowicz, B.P. et al. Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature 392, 608–611 (1998).
Skarnes, W.C. et al. A public gene trap resource for mouse functional genomics. Nat. Genet. 36, 543–544 (2004).
Ivics, Z., Izsvak, Z., Chapman, K.M. & Hamra, F.K. Sleeping Beauty transposon mutagenesis of the rat genome in spermatogonial stem cells. Methods 53, 356–365 (2010).
Chapman, K.M. et al. Rat spermatogonial stem cell mediated gene transfer. in Advanced Protocols for Animal Transgenesis Vol. XVI (eds. Pease, S. & Saunders, T.L.) (Humana Press, 2011).
Grabundzija, I. et al. Comparative analysis of transposable element vector systems in human cells. Mol. Ther. 18, 1200–1209 (2010).
Richardson, T.E., Chapman, K.M., Tenenhaus Dann, C., Hammer, R.E. & Hamra, F.K. Sterile testis complementation with spermatogonial lines restores fertility to DAZL-deficient rats and maximizes donor germline transmission. PLoS One 4, e6308 (2009).
Wu, Z. et al. Spermatogonial culture medium: an effective and efficient nutrient mixture for culturing rat spermatogonial stem cells. Biol. Reprod. 81, 77–86 (2009).
Hamra, F.K. Gene targeting: enter the rat. Nature 467, 161–163 (2010).
Acknowledgements
Work in the authors' laboratories has been supported by EU FP6 (INTHER) and EU FP7 (PERSIST and InduStem), and grants from the Deutsche Forschungsgemeinschaft SPP1230 'Mechanisms of gene vector entry and persistence', and from the Bundesministerium fur Bildung und Forschung (NGFN-2, NGFNplus—ENGINE). Methods for experimental manipulation of rat spermatogonia in culture and for production of mutant rats using spermatogonia were supported by National Institutes of Health grants R21RR023958 from the National Center for Research Resources and RO1HD036022, RO1HD053889, RO1HD061575 from the National Institute of Child Health and Human Development to F.K. Hamra; and by the Cecil H. & Ida Green Center for Reproductive Biology Sciences at the University of Texas Southwestern Medical Center in Dallas.
Author information
Authors and Affiliations
Contributions
F.K.H. and K.M.C. provided concepts on applications of mutant SSC libraries and the use of DAZL-deficient rats as recipient founders; established methods for in vitro culture, gene delivery, clonal selection and transplantation of rat SSC lines for production of mutant rats; supervised the project; and wrote the paper. G.M. produced and edited the movie of F.K.H. showing spermatogonial transplantation procedure. Z. Ivics and Z. Izsvák established SB gene-trap mutagenesis in cultured cells, provided the concept of applying transposon mutagenesis in SSCs, supervised the project and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Video 1
Injection of transfected spermatogonial stem cells into the seminiferous tubules. (MOV 8220 kb)
Rights and permissions
About this article
Cite this article
Ivics, Z., Izsvák, Z., Medrano, G. et al. Sleeping Beauty transposon mutagenesis in rat spermatogonial stem cells. Nat Protoc 6, 1521–1535 (2011). https://doi.org/10.1038/nprot.2011.378
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.378
This article is cited by
-
Long Evans rat spermatogonial lines are effective germline vectors for transgenic rat production
Transgenic Research (2017)
-
Isolation and cultivation of naive-like human pluripotent stem cells based on HERVH expression
Nature Protocols (2016)
-
Germline transgenesis in rodents by pronuclear microinjection of Sleeping Beauty transposons
Nature Protocols (2014)
-
Germline transgenesis in rabbits by pronuclear microinjection of Sleeping Beauty transposons
Nature Protocols (2014)
-
Germline transgenesis in pigs by cytoplasmic microinjection of Sleeping Beauty transposons
Nature Protocols (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.