Abstract
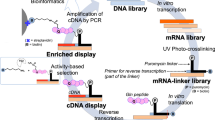
The mRNA display technology enables the in vitro selection and directed evolution of functional proteins from libraries of more than 1012 different mutants in a single test tube. The size of these libraries is well beyond the limit of screening technologies and of most in vivo and in vitro selection methods. The mRNA display technology has been used to select peptides and proteins that bind to a specific ligand, as well as novel enzymes. This protocol details the procedure to produce mRNA-displayed proteins (3 d) and to subject them to a selection and evolution of enzymes for bond-forming reactions (4–10 weeks). This method is demonstrated by the generation of new RNA ligase enzymes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Böttcher, D. & Bornscheuer, U.T. Protein engineering of microbial enzymes. Curr. Opin. Microbiol. 13, 274–282 (2010).
Leemhuis, H., Kelly, R.M. & Dijkhuizen, L. Directed evolution of enzymes: library screening strategies. IUBMB Life 61, 222–228 (2009).
Romero, P.A. & Arnold, F.H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 10, 866–876 (2009).
Jackel, C., Kast, P. & Hilvert, D. Protein design by directed evolution. Ann. Rev. Biophys. 37, 153–173 (2008).
Turner, N.J. Directed evolution drives the next generation of biocatalysts. Nat. Chem. Biol. 5, 568–574 (2009).
Roberts, R.W. & Szostak, J.W. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. USA 94, 12297–12302 (1997).
Nemoto, N., Miyamoto-Sato, E., Husimi, Y. & Yanagawa, H. In vitro virus: Bonding of mRNA bearing puromycin at the 3′-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Lett. 414, 405–408 (1997).
Kurz, M., Gu, K., Al-Gawari, A. & Lohse, P.A. cDNA-Protein fusions: covalent protein-gene conjugates for the in vitro selection of peptides and proteins. Chembiochem. 2, 666–672 (2001).
Takahashi, T.T., Austin, R.J. & Roberts, R.W. mRNA display: ligand discovery, interaction analysis and beyond. Trends Biochem. Sci. 28, 159–165 (2003).
Xu, L.H. et al. Directed evolution of high-affinity antibody mimics using mRNA display. Chem. Biol. 9, 933–942 (2002).
Shen, X.C., Valencia, C.A., Szostak, J., Dong, B. & Liu, R.H. Scanning the human proteome for calmodulin-binding proteins. Proc. Natl. Acad. Sci. USA 102, 5969–5974 (2005).
Fukuda, I. et al. In vitro evolution of single-chain antibodies using mRNA display. Nucleic Acids Res. 34, e127 (2006).
Wilson, D.S., Keefe, A.D. & Szostak, J.W. The use of mRNA display to select high-affinity protein-binding peptides. Proc. Natl. Acad. Sci. USA 98, 3750–3755 (2001).
Raffler, N.A., Schneider-Mergener, J. & Famulok, M. A novel class of small functional peptides that bind and inhibit human alpha-thrombin isolated by mRNA display. Chem. Biol. 10, 69–79 (2003).
Huang, B.C. & Liu, R. Comparison of mRNA-display-based selections using synthetic peptide and natural protein Libraries. Biochemistry 46, 10102–10112 (2007).
Millward, S.W., Takahashi, T.T. & Roberts, R.W. A general route for post-translational cyclization of mRNA display libraries. J. Am. Chem. Soc. 127, 14142–14143 (2005).
Seebeck, F.P. & Szostak, J.W. Ribosomal synthesis of dehydroalanine-containing peptides. J. Am. Chem. Soc. 128, 7150–7151 (2006).
Frankel, A., Millward, S.W. & Roberts, R.W. Encodamers: Unnatural peptide oligomers encoded in RNA. Chem. Biol. 10, 1043–1050 (2003).
Josephson, K., Hartman, M.C.T. & Szostak, J.W. Ribosomal synthesis of unnatural peptides. J. Am. Chem. Soc. 127, 11727–11735 (2005).
Keefe, A.D. & Szostak, J.W. Functional proteins from a random-sequence library. Nature 410, 715–718 (2001).
Liu, R.H., Barrick, J.E., Szostak, J.W. & Roberts, R.W. Optimized synthesis of RNA-protein fusions for in vitro protein selection. Methods Enzymol. 318, 268–293 (2000).
Keefe, A.D. Protein selection using mRNA display. Current Protocols in Molecular Biology 24, 24.5.1–24.5.34 (1 May 2001).
Takahashi, T.T. & Roberts, R.W. In vitro selection of protein and peptide libraries using mRNA display. In Nucleic Acid and Peptide Aptamers: Methods and Protocols - Methods in Molecular Biology Vol. 535 (ed. G. Mayer) (Humana Press, 2009).
Seelig, B. & Szostak, J.W. Selection and evolution of enzymes from a partially randomized non-catalytic scaffold. Nature 448, 828–831 (2007).
Tawfik, D.S. & Griffiths, A.D. Man-made cell-like compartments for molecular evolution. Nat. Biotechnol. 16, 652–656 (1998).
Miller, O.J. et al. Directed evolution by in vitro compartmentalization. Nat. Methods 3, 561–570 (2006).
Hanes, J. & Pluckthun, A. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. USA 94, 4937–4942 (1997).
Zahnd, C., Amstutz, P. & Pluckthun, A. Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat. Methods 4, 269–279 (2007).
Cesaro-Tadic, S. et al. Turnover-based in vitro selection and evolution of biocatalysts from a fully synthetic antibody library. Nat. Biotechnol. 21, 679–685 (2003).
Takahashi, F. et al. Activity-based in vitro selection of T4 DNA ligase. Biochem. Biophys. Res. Commun. 336, 987–993 (2005).
Bloom, J.D. et al. Evolving strategies for enzyme engineering. Curr. Opin. Struct. Biol. 15, 447–452 (2005).
Wang, T.W. et al. Mutant library construction in directed molecular evolution. Mol. Biotechnol. 34, 55–68 (2006).
Shivange, A.V., Marienhagen, J., Mundhada, H., Schenk, A. & Schwaneberg, U. Advances in generating functional diversity for directed protein evolution. Curr. Opin. Chem. Biol. 13, 19–25 (2009).
Reetz, M.T. & Carballeira, J.D. Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nat. Protocols 2, 891–903 (2007).
Cho, G., Keefe, A.D., Liu, R.H., Wilson, D.S. & Szostak, J.W. Constructing high complexity synthetic libraries of long ORFs using in vitro selection. J. Mol. Biol. 297, 309–319 (2000).
Kurz, M., Gu, K. & Lohse, P.A. Psoralen photo-crosslinked mRNA-puromycin conjugates: a novel template for the rapid and facile preparation of mRNA-protein fusions. Nucl. Acids Res. 28, e83 (2000).
Miyamoto-Sato, E. et al. Highly stable and efficient mRNA templates for mRNA-protein fusions and C-terminally labeled proteins. Nucl. Acids Res. 31, e78 (2003).
Sambrook, J. & Russel, D.W. Molecular Cloning: a Laboratory Manual (Cold Spring Harb. Lab. Press, 2001).
Moore, M.J. & Sharp, P.A. Site-specific modification of pre-MRNA: the 2′-hydroxyl groups at the splice sites. Science 256, 992–997 (1992).
Hassur, S.M. & Whitlock, H.W. UV shadowing—a new and convenient method for the location of ultraviolet-absorbing species in polyacrylamide gels. Anal. Biochem. 59, 162–164 (1974).
Sambrook, J. & Russel, D.W. Isolation of DNA fragments from polyacrylamide gels by the crush and soak method. Cold Spring Harb. Protoc. 2006 10.1101/pdb.prot2936 (2006).
Golynskiy, M.V. & Seelig, B. De novo enzymes: from computational design to mRNA display. Trends Biotechnol. 28, 340–345 (2010).
Cadwell, R.C. & Joyce, G.F. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 2, 28–33 (1992).
Wilson, D.S. & Keefe, A.D. Random mutagenesis by PCR. Current Protocols in Molecular Biology Unit 8.3, 8.3.1–8.3.9 2001.
Stemmer, W.P.C. Rapid evolution of a protein in vitro by DNA shuffling. Nature 370, 389–391 (1994).
Wong, T.S., Roccatano, D. & Schwaneberg, U. Steering directed protein evolution: strategies to manage combinatorial complexity of mutant libraries. Environ. Microbiol. 9, 2645–2659 (2007).
Zhao, H.M. & Zha, W.J. In vitro 'sexual' evolution through the PCR-based staggered extension process (StEP). Nat. Protoc. 1, 1865–1871 (2006).
Acknowledgements
The author thanks J.W. Szostak and members of the Szostak lab for their advice and help. The author is supported in part by the National Aeronautics and Space Administration under Agreement No. NNX09AH70A issued through the NASA Astrobiology Institute/Ames Research Center.
Author information
Authors and Affiliations
Contributions
B.S. is solely responsible for this manuscript.
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Rights and permissions
About this article
Cite this article
Seelig, B. mRNA display for the selection and evolution of enzymes from in vitro-translated protein libraries. Nat Protoc 6, 540–552 (2011). https://doi.org/10.1038/nprot.2011.312
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.312
This article is cited by
-
Intracellular directed evolution of proteins from combinatorial libraries based on conditional phage replication
Nature Protocols (2017)
-
Reprogramming eukaryotic translation with ligand-responsive synthetic RNA switches
Nature Methods (2016)
-
In vitro nanobody discovery for integral membrane protein targets
Scientific Reports (2014)
-
Structure and dynamics of a primordial catalytic fold generated by in vitro evolution
Nature Chemical Biology (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.