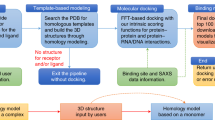
Abstract
Computational docking is the prediction or modeling of the three-dimensional structure of a biomolecular complex, starting from the structures of the individual molecules in their free, unbound form. HADDOCK is a popular docking program that takes a data-driven approach to docking, with support for a wide range of experimental data. Here we present the HADDOCK web server protocol, facilitating the modeling of biomolecular complexes for a wide community. The main web interface is user-friendly, requiring only the structures of the individual components and a list of interacting residues as input. Additional web interfaces allow the more advanced user to exploit the full range of experimental data supported by HADDOCK and to customize the docking process. The HADDOCK server has access to the resources of a dedicated cluster and of the e-NMR GRID infrastructure. Therefore, a typical docking run takes only a few minutes to prepare and a few hours to complete.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
van Dijk, A.D., Boelens, R. & Bonvin, A.M. Data-driven docking for the study of biomolecular complexes. FEBS. J. 272, 293–312 (2005).
Bonvin, A.M. Flexible protein-protein docking. Curr. Opin. Struct. Biol. 16, 194–200 (2006).
Janin, J. Assessing predictions of protein-protein interaction: the CAPRI experiment. Protein Sci. 14, 278–283 (2005).
Méndez, R., Leplae, R., Lensink, M.F. & Wodak, S.J. Assessment of CAPRI predictions in rounds 3-5 shows progress in docking procedures. Proteins 60, 150–169 (2005).
Dominguez, C., Boelens, R. & Bonvin, A.M.J.J. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 (2003).
de Vries, S.J. et al. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins (2007).
van Dijk, A.D.J. et al. Data-driven docking: HADDOCK's adventures in CAPRI. Proteins 60, 232–238 (2005).
de Vries, S.J., van Dijk, A.D.J. & Bonvin, A.M.J.J. WHISCY: What information does surface conservation yield? Application to data-driven docking. Proteins 63, 479–489 (2006).
de Vries, S.J. & Bonvin, A.M. How proteins get in touch: interface prediction in the study of biomolecular complexes. Curr. Protein Pept. Sci. 9, 394–406 (2008).
van Dijk, M. et al. Information-driven protein-DNA docking using HADDOCK: it is a matter of flexibility. Nucleic Acids Res. 34, 3317–3325 (2006).
Berman, H.M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Moreira, I.S., Fernandes, P.A. & Ramos, M.J. Protein-protein docking dealing with the unknown. J. Comput. Chem. (2009).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 (Pt 5): 905–921 (1998).
Schwieters, C.D., Kuszewski, J.J., Tjandra, N. & Clore, G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 (2003).
Vranken, W.F. et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696 (2005).
Hennig, J., Hennig, K.D. & Sunnerhagen, M. MTMDAT: automated analysis and visualization of mass spectrometry data for tertiary and quaternary structure probing of proteins. Bioinformatics 24, 1310–1312 (2008).
Comeau, S.R., Gatchell, D.W., Vajda, S. & Camacho, C.J. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 20, 45–50 (2004).
Tovchigrechko, A. & Vakser, I.A. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 34, W310–314 (2006).
Andrusier, N., Nussinov, R. & Wolfson, H.J. FireDock: fast interaction refinement in molecular docking. Proteins 69, 139–159 (2007).
Mashiach, E. et al. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 36, W229–W232 (2008).
Schneidman-Duhovny, D., Inbar, Y., Nussinov, R. & Wolfson, H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 33, W363–W367 (2005).
Mustard, D. & Ritchie, D.W. Docking essential dynamics eigenstructures. Proteins 60, 269–274 (2005).
Ritchie, D.W. & Kemp, G.J.L. Protein docking using spherical polar Fourier correlations. Proteins 39, 178–194 (2000).
Lyskov, S. & Gray, J.J. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 36, W233–W238 (2008).
Vriend, G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph 8, 52–56, 29 (1990).
Tjandra, N. et al. Use of dipolar H-1-N-15 and H-1-C-13 couplings in the structure determination of magnetically oriented macromolecules in solution. Nat. Struct. Biol. 4, 732–738 (1997).
Meiler, J., Blomberg, N., Nilges, M. & Griesinger, C. A new approach for applying residual dipolar couplings as restraints in structure elucidation. J. Biomol. NMR 16, 245–252 (2000).
Tjandra, N. et al. Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy. Nat. Struct. Biol. 4, 443–449 (1997).
van Dijk, A.D., Fushman, D. & Bonvin, A.M. Various strategies of using residual dipolar couplings in NMR-driven protein docking: application to Lys48-linked di-ubiquitin and validation against 15N-relaxation data. Proteins 60, 367–381 (2005).
van Dijk, A.D., Kaptein, R., Boelens, R. & Bonvin, A.M. Combining NMR relaxation with chemical shift perturbation data to drive protein-protein docking. J. Biomol. NMR 34, 237–244 (2006).
van Dijk, A.D. & Bonvin, A.M. Solvated docking: introducing water into the modelling of biomolecular complexes. Bioinformatics 22, 2340–2347 (2006).
Jmol: an open-source Java viewer for chemical structures in 3D.
Chen, R., Li, L. & Weng, Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins 52, 80–87 (2003).
Wiehe, K. et al. The performance of ZDOCK and ZRANK in rounds 6-11 of CAPRI. Proteins 69, 719–725 (2007).
Schuttelkopf, A.W. & van Aalten, D.M. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta. Crystallogr. D. Biol. Crystallogr. 60, 1355–1363 (2004).
Wang, G. et al. Solution structure of the phosphoryl transfer complex between the signal transducing proteins HPr and IIA(glucose) of the Escherichia coli phosphoenolpyruvate:sugar phosphotransferase system. EMBO J. 19, 5635–5649 (2000).
Hubbard, S.J. & Thornton, J.M. NACCESS (Department of Biochemistry and Molecular biology, University Collage London, 1993).
Linge, J.P., Habeck, M., Rieping, W. & Nilges, M. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics 19, 315–316 (2003).
Nilges, M., Gronenborn, A.M., Brunger, A.T. & Clore, G.M. Determination of three-dimensional structures of proteins by simulated annealing with interproton distance restraints. Application to crambin, potato carboxypeptidase inhibitor and barley serine proteinase inhibitor 2. Protein Eng. 2, 27–38 (1988).
van Dijk, M. & Bonvin, A.M. 3D-DART: a DNA structure modelling server. Nucleic Acids Res. (2009).
Lu, X.J. & Olson, W.K. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 31, 5108–5121 (2003).
Lu, X.J. & Olson, W.K. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 3, 1213–1227 (2008).
Acknowledgements
This work was supported by The Netherlands Organization for Scientific Research (VICI grant no. 700.56.442 to A.B.) and the European Community (FP6 integrated Project SPINE2-COMPLEX, contract no. 032220; FP6 STREP project 'ExtendNMR', contract no. LSHG-CT-2005-018988; and FP7 e-Infrastructure 'e-NMR' I3 project, grant number 213010). The Dutch BiG Grid project with financial support from The Netherlands Organization for Scientific Research (NWO) is acknowledged for the use of the computing and storage facilities. We would like to thank J. Verhaal for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
A.M.J.J.B. is the primary author of the HADDOCK program. S.J.d.V. developed the HADDOCK web server and the Spyder framework. M.v.D. developed the nucleic acid functionality of the server and the graphical design of the server web site. A.M.J.J.B. supervised the project. S.J.d.V. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
de Vries, S., van Dijk, M. & Bonvin, A. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc 5, 883–897 (2010). https://doi.org/10.1038/nprot.2010.32
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2010.32
This article is cited by
-
Identification of WUSCHEL-related homeobox gene and truncated small peptides in transformation efficiency improvement in Eucalyptus
BMC Plant Biology (2023)
-
Visual dynamics: a WEB application for molecular dynamics simulation using GROMACS
BMC Bioinformatics (2023)
-
Employing computational tools to design a multi-epitope vaccine targeting human immunodeficiency virus-1 (HIV-1)
BMC Genomics (2023)
-
Conformational plasticity and allosteric communication networks explain Shelterin protein TPP1 binding to human telomerase
Communications Chemistry (2023)
-
Exploring the viral protease inhibitor space driven by consensus scoring-based virtual screening
In Silico Pharmacology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.