Abstract
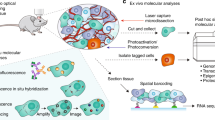
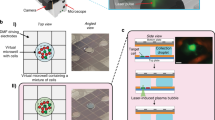
Laser-based microdissection facilitates the isolation of specific cell populations from clinical or animal model tissue specimens for molecular analysis. Expression microdissection (xMD) is a second-generation technology that offers considerable advantages in dissection capabilities; however, until recently the method has not been accessible to investigators. This protocol describes the adaptation of xMD to commonly used laser microdissection instruments and to a commercially available handheld laser device in order to make the technique widely available to the biomedical research community. The method improves dissection speed for many applications by using a targeting probe for cell procurement in place of an operator-based, cell-by-cell selection process. Moreover, xMD can provide improved dissection precision because of the unique characteristics of film activation. The time to complete the protocol is highly dependent on the target cell population and the number of cells needed for subsequent molecular analysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Emmert-Buck, M.R. et al. Laser capture microdissection. Science 274, 998–1001 (1996).
Bonner, R.F. et al. Laser capture microdissection: molecular analysis of tissue. Science 278, 1481–1483 (1997).
Hunt, J.L. & Finkelstein, S.D. Microdissection techniques for molecular testing in surgical pathology. Arch. Pathol. Lab. Med. 128, 1372–1378 (2004).
Espina, V. et al. Laser-capture microdissection. Nat. Protoc. 1, 586–603 (2006).
Dupont Jensen, J. et al. PIK3CA mutations may be discordant between primary and corresponding metastatic disease in breast cancer. Clin. Cancer Res. published online, doi:10.1158/1078-0432.CCR-10-1133 (12 October 2010).
Pinós, T. et al. A novel mutation in the mitochondrial tRNA(Ala) gene (m.5636T>C) in a patient with progressive external ophthalmoplegia. Mitochondrion 11, 228–233 (2011).
Yang, X. et al. Stromal microenvironment processes unveiled by biological component analysis of gene expression in xenograft tumor models. BMC Bioinformatics 11 (Suppl 9), S11 (2010).
Erickson, H.S. et al. Quantitative RT-PCR gene expression analysis of laser microdissected tissue samples. Nat. Protoc. 4, 902–922 (2009).
Johann, D.J. et al. Approaching solid tumor heterogeneity on a cellular basis by tissue proteomics using laser capture microdissection and biological mass spectrometry. J. Proteome. Res. 8, 2310–2318 (2009).
Klein, C.J. et al. Mass spectrometric-based proteomic analysis of amyloid neuropathy type in nerve tissue. Arch. Neurol. published online, doi:10.1001/archneurol.2010.261 (11 October 2010).
Tangrea, M.A. et al. Expression microdissection: operator-independent retrieval of cells for molecular profiling. Diagn. Mol. Pathol. 13, 207–212 (2004).
Hanson, J.A. et al. Gene promoter methylation in prostate tumor-associated stromal cells. J. Natl. Cancer Inst. 98, 255–261 (2006).
Grover, A.C. et al. Tumor-associated endothelial cells display GSTP1 and RARbeta2 promoter methylation in human prostate cancer. J. Transl. Med. 4, 13 (2006).
Goldstein, S.R., McQueen, P.G. & Bonner, R.F. Thermal modeling of laser capture microdissection. Appl. Opt. 37, 7378–7391 (1998).
Perlmutter, M.A. et al. Comparison of snap freezing versus ethanol fixation for gene expression profiling of tissue specimens. J. Mol. Diagn. 6, 371–377 (2004).
Leiva, I.M., Emmert-Buck, M.R. & Gillespie, J.W. Handling of clinical tissue specimens for molecular profiling studies. Curr. Issues Mol. Biol. 5, 27–35 (2003).
Fend, F. et al. Immuno-LCM: laser capture microdissection of immunostained frozen sections for mRNA analysis. Am. J. Pathol. 154, 61–66 (1999).
Rodriguez-Canales, J. et al. Identification of a unique epigenetic sub-microenvironment in prostate cancer. J. Pathol. 211, 410–419 (2007).
Macdonald, J.A., Murugesan, N. & Pachter, J.S. Validation of immuno-laser capture microdissection coupled with quantitative RT-PCR to probe blood-brain barrier gene expression in situ. J. Neurosci. Methods 174, 219–226 (2008).
Liu, Y. et al. Immuno-laser capture microdissection of frozen prolactioma sections to prepare proteomic samples. Colloids Surf. B Biointerfaces 71, 187–193 (2009).
Fend, F., Kremer, M. & Quintanilla-Martinez, L. Laser capture microdissection: methodical aspects and applications with emphasis on immuno-laser capture microdissection. Pathobiology 68, 209–214 (2000).
Eberle, F.C. et al. Immunoguided laser assisted microdissection techniques for DNA methylation analysis of archival tissue specimens. J. Mol. Diagn. 12, 394–401 (2010).
Buckanovich, R.J. et al. Use of immuno-LCM to identify the in situ expression profile of cellular constituents of the tumor microenvironment. Cancer Biol. Ther. 5, 635–642 (2006).
von Smolinski, D., Blessenohl, M., Neubauer, C., Kalies, K. & Gebert, A. Validation of a novel ultra-short immunolabeling method for high-quality mRNA preservation in laser microdissection and real-time reverse transcriptase-polymerase chain reaction. J. Mol. Diagn. 8, 246–253 (2006).
Brown, A.L. & Smith, D.W. Improved RNA preservation for immunolabeling and laser microdissection. RNA 15, 2364–2374 (2009).
Cha, S. et al. In situ proteomic analysis of human breast cancer epithelial cells using laser capture microdissection: annotation by protein set enrichment analysis and gene ontology. Mol. Cell. Proteomics 9, 2529–2544 (2010).
Nonn, L., Vaishnav, A., Gallagher, L. & Gann, P.H. mRNA and micro-RNA expression analysis in laser-capture microdissected prostate biopsies: valuable tool for risk assessment and prevention trials. Exp. Mol. Pathol. 88, 45–51 (2010).
Acknowledgements
This research was supported, in part, by the intramural program of the NIH National Cancer Institute, Center for Cancer Research.
Author information
Authors and Affiliations
Contributions
R.F.B., T.J.P., M.A.T. and M.R.E.-B. developed expression microdissection (xMD). J.C.H., M.A.T. and M.R.E.-B. designed the experiments. J.C.H., S.K., M.D.A. and M.A.T. conducted the tests. J.C.H., J.R.-C., M.A.T. and M.R.E.-B. analyzed the data. J.C.H., M.A.T., J.R.-C. and M.R.E.-B. wrote the manuscript. J.C.H. and M.A.T. contributed equally to the work.
Corresponding author
Ethics declarations
Competing interests
M.E.-B., R.B., and T.P. are inventors on NIH patents covering laser capture microdissection and xMD technology and can receive royalty-based payments through the NIH technology transfer program.
Rights and permissions
About this article
Cite this article
Hanson, J., Tangrea, M., Kim, S. et al. Expression microdissection adapted to commercial laser dissection instruments. Nat Protoc 6, 457–467 (2011). https://doi.org/10.1038/nprot.2010.202
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2010.202
This article is cited by
-
Optimized expression-based microdissection of formalin-fixed lung cancer tissue
Laboratory Investigation (2017)
-
Extensive rewiring of epithelial-stromal co-expression networks in breast cancer
Genome Biology (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.