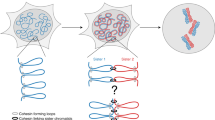
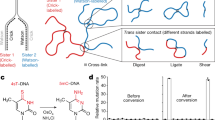
Abstract
Previously, assays for sister chromatid segregation patterns relied on incorporation of 5-bromo-2′-deoxyuridine (BrdU) and indirect methods to infer segregation patterns after two cell divisions. In this study, we describe a method to differentially label sister chromatids of mouse cells and to directly assay sister chromatid segregation patterns after one cell division in vitro and in vivo by adaptation of the well-established CO-FISH technique. BrdU is incorporated into newly formed DNA strands, which are then subjected to photolysis and exonuclease digestion to create single-stranded sister chromatids containing parental template DNA only. Such single-stranded sister chromatids are differentially labeled using unidirectional probes to major satellite sequences coupled to fluorescent markers. Differentially labeled sister chromatids in postmitotic cells are visualized using fluorescence microscopy, and sister chromatid segregation patterns can be directly assayed after one cell division. This procedure requires 4 d for in vivo mouse tissues and 2 d for in vitro–cultured cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lansdorp, P.M. Immortal strands? Give me a break. Cell 129, 1244–1247 (2007).
Rando, T.A. The immortal strand hypothesis: segregation and reconstruction. Cell 129, 1239–1243 (2007).
Falconer, E. et al. Identification of sister chromatids by DNA template strand sequences. Nature 463, 93–97 (2010).
Meyne, J. & Goodwin, E.H. Strand-specific fluorescence in situ hybridization for determining orientation and direction of DNA sequences. Methods Mol. Biol. 33, 141–145 (1994).
Potten, C.S., Hume, W.J., Reid, P. & Cairns, J. The segregation of DNA in epithelial stem cells. Cell 15, 899–906 (1978).
Karpowicz, P. et al. Support for the immortal strand hypothesis: neural stem cells partition DNA asymmetrically in vitro. J. Cell. Biol. 170, 721–732 (2005).
Shinin, V., Gayraud-Morel, B., Gomes, D. & Tajbakhsh, S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat. Cell. Biol. 8, 677–687 (2006).
Smith, G.H. Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands. Development 132, 681–687 (2005).
Cairns, J. Somatic stem cells and the kinetics of mutagenesis and carcinogenesis. Proc. Natl. Acad. Sci. USA 99, 10567–10570 (2002).
Goodwin, E. & Meyne, J. Strand-specific FISH reveals orientation of chromosome 18 alphoid DNA. Cytogenet. Cell Genet. 63, 126–127 (1993).
Bailey, S.M., Goodwin, E.H. & Cornforth, M.N. Strand-specific fluorescence in situ hybridization: the CO-FISH family. Cytogenet. Genome Res. 107, 14–17 (2004).
Goodwin, E.H., Meyne, J., Bailey, S.M. & Quigley, D. On the origin of lateral asymmetry. Chromosoma 104, 345–347 (1996).
Garagna, S. et al. Pericentromeric organization at the fusion point of mouse Robertsonian translocation chromosomes. Proc. Natl. Acad. Sci. USA 98, 171–175 (2001).
Lin, M.S. & Davidson, R.L. Centric fusion, satellite DNA, and DNA polarity in mouse chromosomes. Science 185, 1179–1181 (1974).
Alves, P. & Jonasson, J. New staining method for the detection of sister-chromatid exchanges in BrdU-labelled chromosomes. J. Cell. Sci. 32, 185–195 (1978).
Limoli, C.L. & Ward, J.F. A new method for introducing double-strand breaks into cellular DNA. Radiat. Res. 134, 160–169 (1993).
Lansdorp, P.M. et al. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 5, 685–691 (1996).
Poon, S.S. & Lansdorp, P.M. Quantitative fluorescence in situ hybridization (Q-FISH). Curr. Protoc. Cell Biol. 14 Chapter 18, 18.4.1–18.4.21 (2001).
Horz, W. & Altenburger, W. Nucleotide sequence of mouse satellite DNA. Nucleic Acids Res. 9, 683–696 (1981).
Vissel, B. & Choo, K.H. Mouse major (gamma) satellite DNA is highly conserved and organized into extremely long tandem arrays: implications for recombination between nonhomologous chromosomes. Genomics 5, 407–414 (1989).
Wu, R., Singh, P.B. & Gilbert, D.M. Uncoupling global and fine-tuning replication timing determinants for mouse pericentric heterochromatin. J. Cell. Biol. 174, 185–194 (2006).
Allen, J.W. & Latt, S.A. Analysis of sister chromatid exchange formation in vivo in mouse spermatogonia as a new test system for environmental mutagens. Nature 260, 449–451 (1976).
Poon, S.S., Martens, U.M., Ward, R.K. & Lansdorp, P.M. Telomere length measurements using digital fluorescence microscopy. Cytometry 36, 267–278 (1999).
Gertsenstein, M., Lobe, C. & Nagy, A. ES cell-mediated conditional transgenesis. Methods Mol. Biol. 185, 285–307 (2002).
Acknowledgements
We thank S. Poon for helping proofread the paper. The authors also thank S. Selig, T. de Lange and R.C. Wang for help with CO-FISH protocols. The original study was funded in part by grants from the Canadian Institutes of Health Research (RMF-92093), the Michael Smith Foundation for Health Research, the Canadian Cancer Society Research Institute and the Terry Fox Foundation. We are grateful to the British Columbia Cancer Agency, the Canada Foundation for Innovation, the British Columbia Knowledge Development Fund, the British Columbia Cancer Foundation, the Blusson Fund of the University of British Columbia and the Mahon family for funding the infrastructure that enabled this work.
Author information
Authors and Affiliations
Contributions
E.F. wrote the paper and created the figures; E.C. performed most of the CO-FISH procedure and image acquisition for the original study and proofread the paper; A.H. performed the animal experiments in the original study and proofread the paper; P.M.L. originally developed this procedure, verified the protocol and helped write this paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Falconer, E., Chavez, E., Henderson, A. et al. Chromosome orientation fluorescence in situ hybridization to study sister chromatid segregation in vivo. Nat Protoc 5, 1362–1377 (2010). https://doi.org/10.1038/nprot.2010.102
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2010.102
This article is cited by
-
Single-cell template strand sequencing by Strand-seq enables the characterization of individual homologs
Nature Protocols (2017)
-
Balancing self-renewal against genome preservation in stem cells: How do they manage to have the cake and eat it too?
Cellular and Molecular Life Sciences (2016)
-
A CO-FISH assay to assess sister chromatid segregation patterns in mitosis of mouse embryonic stem cells
Chromosome Research (2013)
-
Evolution of the structure and composition of house mouse satellite DNA sequences in the subgenus Mus (Rodentia: Muridea): a cytogenomic approach
Chromosoma (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.