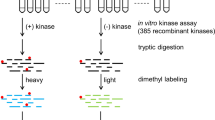
Abstract
Herein, we describe a protocol for the global identification of in vitro substrates targeted by protein kinases using protein microarray technology. Large numbers of fusion proteins tagged at their carboxy-termini are purified in 96-well format and spotted in duplicate onto amino-silane-coated slides in a spatially addressable manner. These arrays are incubated in the presence of purified kinase and radiolabeled ATP, and then washed, dried and analyzed by autoradiography. The extent of phosphorylation of each spot is quantified and normalized, and proteins that are reproducibly phosphorylated in the presence of the active kinase relative to control slides are scored as positive substrates. This approach enables the rapid determination of kinase–substrate relationship on a proteome-wide scale, and although developed using yeast, has since been adapted to higher eukaryotic systems. Expression, purification and printing of the yeast proteome require about 3 weeks. Afterwards, each kinase assay takes ∼3 h to perform.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Hunter, T. Signaling–2000 and beyond. Cell 100, 113 (2000).
Cohen, P. The regulation of protein function by multisite phosphorylation–a 25 year update. Trends Biochem. Sci. 25, 596 (2000).
Elphick, L.M., Lee, S.E., Gouverneur, V. & Mann, D.J. Using chemical genetics and ATP analogues to dissect protein kinase function. ACS Chem. Biol. 2, 299 (2007).
Moffat, J. & Sabatini, D.M. Building mammalian signalling pathways with RNAi screens. Nat. Rev. Mol. Cell Biol. 7, 177 (2006).
Ptacek, J. et al. Global analysis of protein phosphorylation in yeast. Nature 438, 679 (2005).
Schmelzle, K. & White, F.M. Phosphoproteomic approaches to elucidate cellular signaling networks. Curr. Opin. Biotechnol. 17, 406 (2006).
Ptacek, J. & Snyder, M. Charging it up: global analysis of protein phosphorylation. Trends Genet. 22, 545 (2006).
Mok, J. & Snyder, M. in Handbook of Cell Signaling (2009)(in press).
Nita-Lazar, A., Saito-Benz, H. & White, F.M. Quantitative phosphoproteomics by mass spectrometry: past, present, and future. Proteomics 8, 4433 (2008).
Ubersax, J.A. et al. Targets of the cyclin-dependent kinase Cdk1. Nature 425, 859 (2003).
Loog, M. & Morgan, D.O. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 434, 104 (2005).
Dephoure, N. et al. Combining chemical genetics and proteomics to identify protein kinase substrates. Proc. Natl. Acad. Sci. USA 102, 17940 (2005).
Hutti, J.E. et al. A rapid method for determining protein kinase phosphorylation specificity. Nat. Methods 1, 27 (2004).
MacBeath, G. & Schreiber, S.L. Printing proteins as microarrays for high-throughput function determination. Science 289, 1760 (2000).
Zhu, H. et al. Analysis of yeast protein kinases using protein chips. Nat. Genet. 26, 283 (2000).
Gelperin, D.M. et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 19, 2816 (2005).
Brandner, C.J. et al. The ORFeome of Staphylococcus aureus v 1.1. BMC Genomics 9, 321 (2008).
Dricot, A. et al. Generation of the Brucella melitensis ORFeome version 1.1. Genome Res. 14, 2201 (2004).
Lamesch, P. et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics 89, 307 (2007).
Lamesch, P. et al. C. elegans ORFeome version 3.1: increasing the coverage of ORFeome resources with improved gene predictions. Genome Res. 14, 2064 (2004).
Popescu, S.C. et al. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23, 80 (2009).
Popescu, S.C. et al. Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. USA 104, 4730 (2007).
Boyle, S.N. et al. A critical role for cortactin phosphorylation by Abl-family kinases in PDGF-induced dorsal-wave formation. Curr. Biol. 17, 445 (2007).
Cheung, K.H. et al. A dynamic approach to mapping coordinates between microplates and microarrays. J. Biomed. Inform. 35, 306 (2002).
Hastie, C.J., McLauchlan, H.J. & Cohen, P. Assay of protein kinases using radiolabeled ATP: a protocol. Nat. Protoc. 1, 968 (2006).
Zhai, J.Y. Making GenPix Array List (GAL) Files. (2001) (http://www.moleculardevices.com/product_literature/download_doc.php?doc_num=443).
Zhu, X., Gerstein, M. & Snyder, M. ProCAT: a data analysis approach for protein microarrays. Genome Biol. 7, R110 (2006).
Acknowledgements
This work was supported by a grant from the NIH. The assays shown in Figure 1 were performed by Dr. Jason Ptacek and Dr. Geeta Devgan. We thank Dr. Rui Chen and Dr. Vincent Bruno for helpful comments.
Author information
Authors and Affiliations
Contributions
All authors contributed extensively to the work presented in this paper.
Corresponding author
Rights and permissions
About this article
Cite this article
Mok, J., Im, H. & Snyder, M. Global identification of protein kinase substrates by protein microarray analysis. Nat Protoc 4, 1820–1827 (2009). https://doi.org/10.1038/nprot.2009.194
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2009.194
This article is cited by
-
FLEXIQinase, a mass spectrometry–based assay, to unveil multikinase mechanisms
Nature Methods (2012)
-
Enhanced Methylarginine Characterization by Post-Translational Modification-Specific Targeted Data Acquisition and Electron-Transfer Dissociation Mass Spectrometry
Journal of the American Society for Mass Spectrometry (2012)
-
A proteomic approach for the identification of novel lysine methyltransferase substrates
Epigenetics & Chromatin (2011)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.