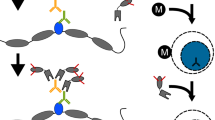
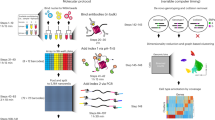
Abstract
Chromatin immunoprecipitation (ChIP) is routinely used to examine epigenetic modification of histones at specific genomic locations. However, covalent modifications of histone tails can serve as docking sites for chromatin regulatory factors. As such, association of these regulatory factors with chromatin could cause steric hindrance for antibody recognition, resulting in an underestimation of the relative enrichment of a given histone modification at specific loci. To overcome this problem, we have developed a native ChIP protocol to study covalent modification of histones that takes advantage of hydroxyapatite (HAP) chromatography to wash away chromatin-associated proteins before the immunoprecipitation of nucleosomes. This fast and simple procedure consists of five steps: nuclei isolation from cultured cells; fragmentation of chromatin using MNase; purification of nucleosomes using HAP; immunoprecipitation of modified nucleosomes; and qPCR analysis of DNA associated with modified histones. Nucleosomes prepared in this manner are free of contaminating proteins and permit an accurate evaluation of relative abundance of different covalent histone modifications at specific genomic loci. Completion of this protocol requires ∼1.5 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hebbes, T.R., Thorne, A.W. & Crane-Robinson, C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 7, 1395–1402 (1988).
O'Neill, L.P. & Turner, B.M. Immunoprecipitation of native chromatin: NChIP. Methods 31, 76–82 (2003).
O'Neill, L.P., VerMilyea, M.D. & Turner, B.M. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat. Genet. 38, 835–841 (2006).
Weber, M. et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 37, 853–862 (2005).
Dorbic, T. & Wittig, B. Isolation of oligonucleosomes from active chromatin using HMG17-specific monoclonal antibodies. Nucleic Acids Res. 14, 3363–3376 (1986).
Solomon, M.J., Larsen, P.L. & Varshavsky, A. Mapping protein–DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell 53, 937–947 (1988).
Orlando, V., Strutt, H. & Paro, R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11, 205–214 (1997).
Dhalluin, C. et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399, 491–496 (1999).
Santos-Rosa, H. et al. Methylation of histone H3 K4 mediates association of the isw1p ATPase with chromatin. Mol. Cell 12, 1325–1332 (2003).
Wysocka, J. et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442, 86–90 (2006).
Strahl, B.D. & Allis, C.D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Ruthenburg, A.J., Allis, C.D. & Wysocka, J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 25, 15–30 (2007).
Vermeulen, M. et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69 (2007).
Bernardi, G. Chromatography of nucleic acids on hydroxyapatite. Nature 206, 779–783 (1965).
Thorne, A.W., Cary, P.D. & Crane-Robinson, C. Extraction and separation of core histone and non-histone chromosomal proteins. in Chromatin: A Practical Approach Vol. 9 (ed. Gould, H.) 35–57 (Oxford University Press, Oxford, 1998).
Rampalli, S. et al. p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat. Struct. Mol. Biol. 14, 1150–1156 (2007).
Ren, B. et al. Genome-wide location and function of DNA binding proteins. Science 290, 2306–2309 (2000).
Lee, T.I., Johnstone, S.E. & Young, R.A. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1, 729–748 (2006).
Mikkelsen, T.S. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007).
Hager, G.L. & Fragoso, G. Analysis of nucleosome positioning in mammalian cells. Methods Enzymol. 304, 626–638 (1999).
Lomvardas, S. & Thanos, D. Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110, 261–271 (2002).
Sambrook, J. & Russell, D.W. Molecular Cloning: A Laboratory Manual (Cold Spring Laboratory Press, New York, 2001).
Acknowledgements
This work was supported by grants from the Canadian Institutes of Health Research, Muscular Dystrophy Association and the Stem Cell Network (to F.J.D.); and Terry Fox Foundation/National Cancer Institute of Canada, and Human Frontiers Science Program (to M.B.). F.J.D. holds a Canadian Research Chair in Epigenetic Regulation of Transcription. M.B. holds a Canadian Research Chair in the Regulation of Gene Expression.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Brand, M., Rampalli, S., Chaturvedi, CP. et al. Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation of nucleosomes isolated using hydroxyapatite chromatography. Nat Protoc 3, 398–409 (2008). https://doi.org/10.1038/nprot.2008.8
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2008.8
This article is cited by
-
Epigenetic malleability at core promoter initiates tobacco PR-1a expression post salicylic acid treatment
Molecular Biology Reports (2023)
-
5-Hydroxymethylcytosine (5hmC) at or near cancer mutation hot spots as potential targets for early cancer detection
BMC Research Notes (2022)
-
HIRA stabilizes skeletal muscle lineage identity
Nature Communications (2021)
-
A native chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in strawberry fruits
Plant Methods (2020)
-
Histone deacetylase (HDAC) 1 and 2 complexes regulate both histone acetylation and crotonylation in vivo
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.