Abstract
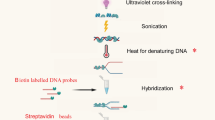
Chromatin immunoprecipitation (ChIP) is a powerful tool for the characterization of covalent histone modifications and DNA–histone interactions in vivo. The procedure includes DNA–histone cross-linking in chromatin, shearing DNA into smaller fragments, immunoprecipitation with antibodies against the histone modifications of interest, followed by PCR identification of associated DNA sequences. In this protocol, we describe a simplified and optimized version of ChIP assay by reducing the number of experimental steps and isolation solutions and shortening preparation times. We include a nuclear isolation step before chromatin shearing, which provides a good yield of high-quality DNA resulting in at least 15 μg of DNA from each immunoprecipitated sample (from 0.2 to 0.4 g of starting tissue material) sufficient to test ≥25 genes of interest. This simpler and cost-efficient protocol has been applied for histone-modification studies of various Arabidopsis thaliana tissues and is easy to adapt for other systems as well.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Van Driel, R., Fransz, P.F. & Verschure, P.J. The eukaryotic genome: a system regulated at different hierarchical levels. J. Cell Sci. 15, 4067–4075 (2003).
Fried, M.G. Measurement of protein-DNA interaction parameters by electrophoresis mobility shift assay. Electrophoresis 10, 366–376 (1989).
Hellman, L.M. & Fried, M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein–nucleic acid interactions. Nat. Protoc. 2, 1849–1861 (2007).
Wood, K.V. Marker proteins for gene expression. Curr. Opin. Biotechnol. 6, 50–58 (1995).
Chalfie, M., Tu, Y., Euskirchen, G., Ward, W.W. & Prasher, D.C. Green fluorescent protein as a marker for gene expression. Science 263, 802–805 (1994).
Bulyk, L.M. DNA microarray technologies for measuring protein–DNA interactions. Curr. Opin. Biotechnol. 17, 422–430 (2006).
Bonaldi, T., Regula, J.T. & Imhof, A. The use of mass spectrometry for the analysis of histone modifications. Methods Enzymol. 377, 111–130 (2004).
Burlingame, A.L., Zhang, X. & Chalkley, R.J. Mass spectrometric analysis of histone posttranslational modifications. Methods 36, 383–394 (2005).
Wang, M.M. & Reed, R.R. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature 364, 121–126 (1993).
Feng, S.Y., Ota, K., Yamada, Y., Sawabu, N. & Ito, T. A yeast one-hybrid system to detect methylation-dependent DNA-protein interactions. Biochem. Biophys. Res. Commun. 313, 922–925 (2004).
Orlando, V., Strutt, H. & Paro, R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11, 205–214 (1997).
Orlando, V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 25, 99–104 (2000).
Kuo, M.H. & Allis, C.D. In vivo cross-linking and immunoprecipitation for studying dynamic Protein: DNA associations in a chromatin environment. Methods 19, 425–433 (1999).
Huebert, D.J., Kamal, M., O'Donovan, A. & Bernstein, B.E. Genome-wide analysis of histone modifications by ChIP-on-chip. Methods 40, 365–369 (2006).
Nelson, J.D., Denisenko, O. & Bomsztyk, K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1, 179–185 (2006).
Alvarez-Venegas, R. & Avramova, Z. Methylation patterns of histone H3 Lys 4, Lys 9 and Lys 27 in transcriptionally active and inactive Arabidopsis genes and in atx1 mutants. Nucleic Acids Res. 33, 5199–5207 (2005).
Chaya, D. & Zaret, K.S. Sequential chromatin immunoprecipitation from animal tissues. Methods Enzymol. 376, 361–372 (2004).
Ezhkova, E. & Tansey, W.P. Chromatin immunoprecipitation to study protein-DNA interactions in budding yeast. Methods Mol. Biol. 313, 225–244 (2006).
Sandmann, T., Jakobsen, J.S. & Furlong, EE. ChIP-on-chip protocol for genome-wide analysis of transcription factor binding in Drosophila melanogaster embryos. Nat. Protoc. 1, 2839–2855 (2006).
Ascenzi, R. & Gantt, J.S. Subnuclear distribution of the entire complement of linker histone variants in Arabidopsis thaliana. Chromosoma 108, 345–355 (1999).
Marty, F. Plant vacuoles. Plant Cell 11, 587–600 (1999).
Reisen, D., Marty, F. & Leborgne-Castel, N. New insights into the tonoplast architecture of plant vacuoles and vacuolar dynamics during osmotic stress. BMC Plant Biol. 5, 13 (2005).
Müntz, K. Protein dynamics and proteolysis in plant vacuoles. J. Exp. Botany 58, 2391–2407 (2007).
Jackson, J.P. et al. Dimethylation of histone H3 Lys 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112, 308–315 (2004).
Bowler, C. et al. Chromatin techniques for plant cells. Plant J. 39, 776–789 (2004).
Gendrel, A.V., Lippman, Z., Martienssen, R. & Colot, V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2, 213–218 (2005).
Gendrel, A.V., Lippman, Z., Yordan, C., Colot, V. & Martienssen, R.A. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297, 1871–1873 (2002).
Johnson, M., Cao, X. & Jacobsen, S. Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12, 1360–1367 (2002).
Haring, M. et al. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Meth. 3, 11 (2007).
Zhang, X. et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5, 1026–1035 (2007).
Turck, F. et al. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3, e86 (2007).
Alvarez-Venegas, R., Abdallat, A.A., Guo, M., Alfano, J. & Avramova, Z. Epigenetic control of transcription factor at the cross section of two antagonistic pathways. Epigenetics 2, 106–117 (2007).
Saleh, A., Al-Abdallat, A., Ndamukong, I., Alvarez-Venegas, R. & Avramova, Z. The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish “bivalent chromatin marks” at the silent AGAMOUS locus. Nucleic Acids Res. 35, 6290–6296 (2007).
Saleh, A. et al. The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode divergent biochemical functions. Plant Cell (in press) 10.1105/tpc.107.056614 (2008).
Takada, S. & Goto, K. Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15, 2856–2865 (2003).
Teper-Bamnolkerm, P. & Samach, A. The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17, 2661–2675 (2005).
Cao, R. & Zhang, Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 14, 155–164 (2004).
Goodrich, J. et al. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386, 44–51 (1997).
Schubert, D., Clarenz, O. & Goodrich, J. Epigenetic control of plant development by Polycomb-group proteins. Curr. Opin. Plant Biol. 8, 553–561 (2005).
Chanvivattana, Y. et al. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131, 5263–5276 (2004).
Jack, T. Molecular and genetic mechanisms of floral control. Plant Cell 16, S1–S17 (2004).
Acknowledgements
The authors are grateful to Dr Malali Gowda for critically reading the manuscript and Dr Justin Goodrich for his gift of clf mutant seeds. This work was partially supported by the NSF grant MCB-0343934 to Z.A.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Saleh, A., Alvarez-Venegas, R. & Avramova, Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3, 1018–1025 (2008). https://doi.org/10.1038/nprot.2008.66
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2008.66
This article is cited by
-
Rewiring of a KNOXI regulatory network mediated by UFO underlies the compound leaf development in Medicago truncatula
Nature Communications (2024)
-
Control of arbuscule development by a transcriptional negative feedback loop in Medicago
Nature Communications (2023)
-
Epigenetic silencing of callose synthase by VIL1 promotes bud-growth transition in lily bulbs
Nature Plants (2023)
-
Molecular basis of methyl-salicylate-mediated plant airborne defence
Nature (2023)
-
Comprehensive molecular evaluation of the histone methyltransferase gene family and their important roles in two-line hybrid wheat
BMC Plant Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.