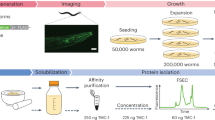
Abstract
It is often difficult to produce eukaryotic membrane proteins in large quantities, which is a major obstacle for analyzing their biochemical and structural features. To date, yeast has been the most successful heterologous overexpression system in producing eukaryotic membrane proteins for high-resolution structural studies. For this reason, we have developed a protocol for rapidly screening and purifying eukaryotic membrane proteins in the yeast Saccharomyces cerevisiae. Using this protocol, in 1 week many genes can be rapidly cloned by homologous recombination into a 2 μ GFP-fusion vector and their overexpression potential determined using whole-cell and in-gel fluorescence. The quality of the overproduced eukaryotic membrane protein-GFP fusions can then be evaluated over several days using confocal microscopy and fluorescence size-exclusion chromatography (FSEC). This protocol also details the purification of targets that pass our quality criteria, and can be scaled up for a large number of eukaryotic membrane proteins in either an academic, structural genomics or commercial environment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Drew, D.E., von Heijne, G., Nordlund, P. & de Gier, J.W. Green fluorescent protein as an indicator to monitor membrane protein overexpression in Escherichia coli. FEBS Lett. 507, 220–224 (2001).
Drew, D., Lerch, M., Kunji, E., Slotboom, D.J. & de Gier, J.W. Optimization of membrane protein overexpression and purification using GFP fusions. Nat. Methods 3, 303–313 (2006).
Drew, D. et al. A scalable, GFP-based pipeline for membrane protein overexpression screening and purification. Protein Sci. 14, 2011–2017 (2005).
Kawate, T. & Gouaux, E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006).
Jasti, J., Furukawa, H., Gonzales, E.B. & Gouaux, E. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature 449, 316–323 (2007).
Newstead, S., Kim, H., von Heijne, G., Iwata, S. & Drew, D. High-throughput fluorescent-based optimization of eukaryotic membrane protein overexpression and purification in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 104, 13936–13941 (2007).
Ferguson, A.D. et al. Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein. Science 317, 510–512 (2007).
Nyblom, M. et al. Exceptional overproduction of a functional human membrane protein. Protein Expr. Purif. 56, 110–120 (2007).
Jidenko, M. et al. Crystallization of a mammalian membrane protein overexpressed in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 102, 11687–11691 (2005).
Pedersen, B.P., Buch-Pedersen, M.J., Morth, J.P., Palmgren, M.G. & Nissen, P. Crystal structure of the plasma membrane proton pump. Nature 450, 1111–1114 (2007).
Ellgaard, L. & Helenius, A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell. Biol. 4, 181–191 (2003).
Nakatsukasa, K., Huyer, G., Michaelis, S. & Brodsky, J.L. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell 132, 101–112 (2008).
Niebauer, R.T. & Robinson, A.S. Exceptional total and functional yields of the human adenosine (A2a) receptor expressed in the yeast Saccharomyces cerevisiae. Protein Expr. Purif. 46, 204–211 (2006).
Kota, J., Gilstring, C.F. & Ljungdahl, P.O. Membrane chaperone Shr3 assists in folding amino acid permeases preventing precocious ERAD. J. Cell Biol. 176, 617–628 (2007).
Cormack, B.P. et al. Yeast-enhanced green fluorescent protein (yEGFP)a reporter of gene expression in Candida albicans. Microbiology 143 (Pt 2), 303–311 (1997).
Lucast, L.J., Batey, R.T. & Doudna, J.A. Large-scale purification of a stable form of recombinant tobacco etch virus protease. Biotechniques 30, 544–546, 548, 550 passim (2001).
Mumberg, D., Müller, R. & Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122 (1995).
Sato, K., Sato, M. & Nakano, A. Rer1p, a retrieval receptor for endoplasmic reticulum membrane proteins, is dynamically localized to the Golgi apparatus by coatomer. J. Cell Biol. 152, 935–944 (2001).
Wagner, S., Bader, M.L., Drew, D. & de Gier, J.W. Rationalizing membrane protein overexpression. Trends Biotechnol. 24, 364–371 (2006).
André, N. et al. Enhancing functional production of G protein-coupled receptors in Pichia pastoris to levels required for structural studies via a single expression screen. Protein Sci. 15, 1115–1126 (2006).
Drew, D. et al. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc. Natl. Acad. Sci. USA 99, 2690–2695 (2002).
Huh, W.K. et al. Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003).
Guan, L., Mirza, O., Verner, G., Iwata, S. & Kaback, H.R. Structural determination of wild-type lactose permease. Proc. Natl. Acad. Sci. USA 104, 15294–15298 (2007).
Long, S.B., Tao, X., Campbell, E.B. & MacKinnon, R. Atomic structure of a voltage-dependent K· channel in a lipid membrane-like environment. Nature 450, 376–382 (2007).
Burke, D., Dawson, D. & Stearns, T. Methods in Yeast Genetics, Cold Spring Harbor Laboratory Course Manual, 2000 edition (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2000).
Stimpson, H.E., Lewis, M.J. & Pelham, H.R. Transferrin receptor-like proteins control the degradation of a yeast metal transporter. EMBO J. 25, 662–672 (2006).
Galan, J.M., Moreau, V., Andre, B., Volland, C. & Haguenauer-Tsapis, R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J. Biol. Chem. 271, 10946–10952 (1996).
Liu, J., Sitaram, A. & Burd, C.G. Regulation of copper-dependent endocytosis and vacuolar degradation of the yeast copper transporter, Ctr1p, by the Rsp5 ubiquitin ligase. Traffic 8, 1375–1384 (2007).
Acknowledgements
We thank Jan-Willem de Gier, Mitsunori Shiroishi, Shuichiro Goda and the reviewers for critically reading the manuscript and for useful comments. D.D. was a recipient of an European Molecular Biology Organization (EMBO) long-term fellowship. Funded by grants from the European Membrane Protein Consortium (E-MEP), the Membrane Protein Structure Initiative (MPSI) and the Biotechnology and Biological Sciences Research Council (BBSRC) (to S.I.). The project was also supported by grant number R01GM081827 from the National Institute of General Medical Sciences (to G.v.H. and H.K.); the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Drew, D., Newstead, S., Sonoda, Y. et al. GFP-based optimization scheme for the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae. Nat Protoc 3, 784–798 (2008). https://doi.org/10.1038/nprot.2008.44
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2008.44
This article is cited by
-
TMEM16 scramblases thin the membrane to enable lipid scrambling
Nature Communications (2022)
-
Fluorescence-detection size-exclusion chromatography utilizing nanobody technology for expression screening of membrane proteins
Communications Biology (2021)
-
The GFP thermal shift assay for screening ligand and lipid interactions to solute carrier transporters
Nature Protocols (2021)
-
The molecular basis for sugar import in malaria parasites
Nature (2020)
-
Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion
Nature Structural & Molecular Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.