Abstract
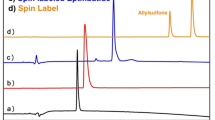
In this protocol, we describe the facile synthesis of the nitroxide spin-label 2,2,5,5-tetramethyl-pyrrolin-1-oxyl-3-acetylene (TPA) and then its coupling to DNA/RNA through Sonogashira cross-coupling during automated solid-phase synthesis. Subsequently, we explain how to perform distance measurements between two such spin-labels on RNA/DNA using the pulsed electron paramagnetic resonance method pulsed electron double resonance (PELDOR). This combination of methods can be used to study global structure elements of oligonucleotides in frozen solution at RNA/DNA amounts of ∼10 nmol. We especially focus on the Sonogashira cross-coupling step, the advantages of the ACE chemistry together with the appropriate parameters for the RNA synthesizer and on the PELDOR data analysis. This procedure is applicable to RNA/DNA strands of up to ∼80 bases in length and PELDOR yields reliably spin–spin distances up to ∼6.5 nm. The synthesis of TPA takes ∼5 days and spin labeling together with purification ∼4 days. The PELDOR measurements usually take ∼16 h and data analysis from an hour up to several days depending on the extent of analysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Berliner, L.J. (ed.) Biological Magnetic Resonance. Spin Labeling. The Next Millennium. Vol. 14 (Plenum Press, New York, USA, 1998).
Caron, M. & Dugas, H. Specific spin-labeling of transfer ribonucleic acid molecules. Nuceic Acids Res. 3, 19–34 (1976).
Luoma, G.A., Herring, F.G. & Marshall, A.G. Flexibility of end-labeled polymers from electron spin resonance line-shape analysis: 3′ terminus of transfer ribonucleic acid and 5S ribonucleic acid. Biochemistry 21, 6591–6598 (1982).
Hara, H., Horiuche, T., Saneyoshi, M. & Nishimura, S. 4-Thiouridine-specific spin-labeling. Biochem. Biophys. Res. Commun. 38, 305–311 (1970).
McIntosh, A.R., Caron, M. & Dugas, H. A specific spin labeling of the anticodon of E. coli tRNAGlu . Biochem. Biophys. Res. Commun. 55, 1356–1362 (1973).
Nagahara, S., Murakami, A. & Makino, K. Spin-labeled oligonucleotides site specifically labeled at the internucleotide linkage. Separation of stereoisomeric probes and EPR spectroscopical detection of hybrid formation in solution. Nucleosides Nucleotides 11, 889–901 (1992).
Qin, P.Z., Butcher, S.E., Feigon, J. & Hubbell, W.L. Quantitative analysis of the isolated GAAA tetraloop/receptor interaction in solution: a site-directed spin labeling study. Biochemistry 40, 6929–6936 (2001).
Cai, Q. et al. Site-directed spin labeling measurements of nanometer distances in nucleic acids using a sequence-independent nitroxide probe. Nucleic Acids Res. 34, 4722–4730 (2006).
Edwards, T.E., Okonogi, T.M., Robinson, B.H. & Sigurdsson, S.T. Site-specific incorporation of nitroxide spin-labels into internal sites of the TAR RNA; structure-dependent dynamics of RNA by EPR spectroscopy. J. Am. Chem. Soc. 123, 1527–1528 (2001).
Kim, N.-K., Murali, A. & DeRose, V.J. A distance ruler for RNA using EPR and site-directed spin labeling. Chem. Biol. 11, 939–948 (2004).
Qin, P.Z., Hideg, K., Feigon, J. & Hubbell, W.L. Monitoring RNA base structure and dynamics using site-directed spin labeling. Biochemstry 42, 6772–6783 (2003).
Ramos, A. & Varani, G. A new method to detect long-range protein–RNA contacts: NMR detection of electron–proton relaxation induced by nitroxide spin-labeled RNA. J. Am. Chem. Soc. 120, 10992–10993 (1998).
Spaltenstein, A., Robinson, B.H. & Hopkins, P.B. A rigid and non-perturbing probe for duplex DNA motion. J. Am. Chem. Soc. 110, 1299–1301 (1988).
Spaltenstein, A., Robinson, B.H. & Hopkins, P.B. Sequence- and structure-dependent DNA base dynamics: synthesis, structure, and dynamics of site and sequence specifically spin-labeled DNA. Biochemistry 28, 9484–9495 (1989).
Hustedt, E.J., Kirchner, J.J., Spaltenstein, A., Hopkins, P.B. & Robinson, B.H. Monitoring DNA dynamics using spin-labels with different independent mobilities. Biochemistry 34, 4369–4375 (1995).
Gannett, P.M. et al. Probing triplex formation by EPR spectroscopy using a newly synthesized spin label for oligonucleotides. Nucleic Acids Res. 30, 5328–5337 (2002).
Schiemann, O., Piton, N., Mu, Y., Stock, G., Engels, J.W. & Prisner, T.F. A PELDOR-based nanometer distance ruler for oligonucleotides. J. Am. Chem. Soc. 126, 5722–5729 (2004).
Piton, N., Schiemann, O., Mu, Y., Stock, G., Prisner, T.F. & Engels, J.W. Synthesis of spin-labeled RNAs for long range distance measurements by PELDOR. Nucleosides Nucleotides Nucleic Acids 24, 771–775 (2005).
Piton, N., Mu, Y., Stock, G., Prisner, T.F., Schiemann, O. & Engels, J.W. Nucleic Acids Res. (in the press).
Miller, T.R. et al. A probe for sequence-dependent nucleic acid dynamics. J. Am. Chem. Soc. 117, 9377–9378 (1995).
Okonogi, T.M., Reese, A.W., Alley, S.C., Hopkins, P.B. & Robinson, B.H. Flexibility of duplex DNA on the sub-microsecond timescale. Biophys. J. 77, 3256–327 (1999).
Barhate, N., Cekan, P., Massey, A.P. & Sigurdssson, S.T. A nucleoside that contains a rigid nitroxide spin label: a fluorophore in disguise. Angew. Chem. Int. Ed. 46, 2655–2658 (2007).
Sprinzl, M., Scheit, K.H. & Cramer, F. Preparation in-vitro of a 2-thiocytidine-containing yeast transfer-RNA PHE-A73-C74-S2C75-A76 and its interaction with para-hydroxymercuribenzoate. Eur. J. Biochem. 34, 306–310 (1973).
Sprinzl, M., Krämer, E. & Stehlik, D. On the structure of phenylalanine tRNA from yeast. Eur. J. Biochem. 49, 595–605 (1974).
Macosko, J.C., Pio, M.S., Tinoco, J.R. & Shin, Y.-K. A novel 5′ displacement spin-labeling technique for electron paramagnetic resonance spectroscopy of RNA. RNA 5, 1158–1166 (1999).
Bobst, A.M., Pauly, G.T., Keyes, R.S. & Bobst, E.V. Enzymatic sequence-specific spin labeling of a DNA fragment containing the recognition sequence of EcoRI endonuclease. FEBS Lett. 228, 33–36 (1988).
Liang, Z., Freed, J.H., Keyes, R.S. & Bobst, A.M. An electron spin resonance study of DNA dynamics using the slowly relaxing local structure model. J. Phys. Chem. B. 104, 5372–5381 (2000).
Keyes, R.S. & Bobst, A.M. Spin-labeled nucleic acids. in Biological Magnetic Resonance. Spin Labeling. The Next Millennium Vol. 14 (ed. Berliner, L.J.) 7.283–7.334 (Plenum Press, New York, 1998).
Okonogi, T.M., Alley, S.C., Reese, A.W., Hopkins, P.B. & Robinson, B.H. Sequence-dependent dynamics in duplex DNA. Biophys. J. 78, 2560–2571 (2000).
van Doorslaer, S. & Jeschke, G. Dynamics by EPR: picosecond to microsecond time scales. in Fluxional Organometallic and Coordination Compounds. (eds. Gielen, M., Willem, R. & Wrackmeyer, B.) 6.219–6.242 (Wiley, Weinheim, 2004).
Jacobsen, K., Hubbell, W.L., Ernst, O.P. & Risse, T. Details of the partial unfolding of T4 lysozyme on quartz using site-directed spin labeling. Angew. Chem. Int. Ed. 45, 3874–3877 (2006).
Potapenko, D.I. et al. Real-time monitoring of drug-induced changes in the stomach acidity of living rats using improved pH-sensitive nitroxides and low-field EPR techniques. J. Magn. Reson. 182, 1–11 (2006).
Steinhoff, H.J. Inter- and intra-molecular distances determined by EPR spectroscopy and site-directed spin labeling reveal protein–protein and protein–oligonucleotide interaction. Biol. Chem. 385, 913–920 (2004).
Halpern, H.J. et al. Diminished aqueous microviscosity of tumors in murine models measured with in vivo radiofrequency electron paramagnetic resonance. Cancer Res. 59, 5836–5841 (1999).
Liang, B.Y., Bushweller, J.H. & Tamm, L.K. Site-directed parallel spin-labeling and paramagnetic relaxation enhancement in structure determination of membrane proteins by solution NMR spectroscopy. J. Am. Chem. Soc. 128, 4389–4397 (2006).
Berliner, L.J., Eaton, S.S. & Eaton, G.R. (eds.) Biological Magnetic Resonance. Distance Measurements in Biological Systems by EPR Vol. 19 (Kluwer Academic, New York, 2000).
Jeschke, G., Bender, A., Paulsen, H., Zimmermann, H. & Godt, A. Sensitivity enhancement in pulse EPR distance measurements. J. Magn. Reson. 169, 1–12 (2004).
Milov, A.D., Salikov, K.M. & Shirov, M.D. Application of the double resonance method to electron spin echo in a study of the spatial distribution of paramagnetic centers in solids. Sov. Phys. Solid State 23, 565–569 (1981).
Martin, R.E. et al. Determination of the end-to-end distances in a series of TEMPO diradicals of up to 2.8 nm length with a new four-pulse double electron electron resonance experiment. Angew. Chem. Int. Ed. 37, 2834–2837 (1998).
Milov, A.D., Maryasov, A.G. & Tsvetkov, Y.D. Pulsed electron double resonance (PELDOR) and its application in free-radicals research. Appl. Magn. Reson. 15, 107–143 (1998).
Mims, W.B. in Electron Paramagnetic Resonance (ed. Geschwind, S.) 263–264 (Plenum Press, New York, 1972).
Weber, A., Schiemann, O., Bode, B. & Prisner, T.F. PELDOR at S- and X-band frequencies and the separation of exchange coupling from dipolar coupling. J. Magn. Reson. 157, 277–285 (2002).
Schiemann, O., Weber, A., Edwards, T.E., Prisner, T.F. & Sigurdsson, S.T. Nanometer distance measurements on RNA using PELDOR. J. Am. Chem. Soc. 125, 3434–3435 (2003).
Milov, A.D. et al. The secondary structure of a membrane-modifying peptide in a supramolecular assembly studied by PELDOR and cw-ESR spectroscopy. J. Am. Chem. Soc. 123, 3784–3789 (2001).
Denysenkov, V.P., Prisner, T.F., Stubbe, J. & Bennati, M. High-field pulsed electron-electron double resonance spectroscopy to determine the orientation of the tyrosyl radicals in ribonucleotide reductase. Proc. Natl. Acad. Sci. USA 103, 13386–13390 (2006).
Elsässer, C., Brecht, M. & Bittl, R. Pulsed electron–electron double resonance on multinuclear metal clusters: assignment of spin projection factors based on the dipolar interaction. J. Am. Chem. Soc. 124, 12606–12611 (2002).
Hara, H., Kawamori, A., Astashkin, A.V. & Ono, T. The distance from tyrosine D to redox-active components on the donor side of photosystem II determined by pulsed electron–electron double resonance. Biochim. Biophys. Acta 1276, 140–146 (1996).
Raitsimring, A. “2+1” Pulse sequence as applied for distance and spatial distribution measurements of paramagnetic centers. in Biological Magnetic Resonance. Distance Measurements in Biological Systems by EPR Vol. 19 (eds. Berliner, L.J., Eaton, S.S. & Eaton, G.R.) 10.461–10.490 (Kluwer Academic, New York, 2000).
Borbat, P.P., Davis, J.H., Butcher, S.E. & Freed, J.H. Measurement of large distances in biomolecules using double quantum filtered refocused electron spin-echoes. J. Am. Chem. Soc. 126, 7746–7747 (2004).
Eaton, S.S. & Eaton, G.R. Determination of distances based on T1 and Tm effects. in Biological Magnetic Resonance. Distance Measurements in Biological Systems by EPRF. Vol. 19 (eds. Berliner, L.J., Eaton, S.S. & Eaton, G.R.) 8.348–8.378 (Kluwer Academic, New York, 2000).
Lakowicz, J.R. Principles of Fluorescence Spectroscopy. 3rd edn., 13.443–13.471 (Springer, New York, 2006).
Merritt, M.E., Sigurdsson, S.T. & Drobny, G.P. Long-range distance measurements to the phosphodiester backbone of solid nucleic acids using P-31-F-19 REDOR NMR. J. Am. Chem. Soc. 121, 6070–6071 (1999).
Scaringe, S.A., Kitchen, D., Kaiser, R. & Marshall, W.S. Preparation of 5′-silyl-2′-orthoester ribonucleosides for use in oligoribonucleotide synthesis. Curr. Prot. Nucleics Acids Chem. 2.10.1–2.10.15 (2004).
Scaringe, S.A. RNA oligonucleotide synthesis via 5′-silyl-2′-orthoester chemistry. Methods 23, 206–217 (2001).
Hideg, K., Hankovszky, H.O., Lex, L. & Kulcsár, G. Nitroxyls: VI. Synthesis and reactions of 3-hydroxymethyl-2,2,5,5-tetramethyl-2,5-dihydropyrrole-1-oxyl and 3-formyl derivatives. Synthesis 911–914 (1980).
Bowman, M.K., Maryasov, A.G., Kim, N.-K. & DeRose, V.J. Visualization of distance distribution from pulsed double electron–electron resonance data. Appl. Magn. Res. 26, 23–29 (2004).
Acknowledgements
We thank D. Margraf for helpful advice on this manuscript. This work was supported by the DFG within the SFB 579.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Schiemann, O., Piton, N., Plackmeyer, J. et al. Spin labeling of oligonucleotides with the nitroxide TPA and use of PELDOR, a pulse EPR method, to measure intramolecular distances. Nat Protoc 2, 904–923 (2007). https://doi.org/10.1038/nprot.2007.97
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.97
This article is cited by
-
In situ observation of conformational dynamics and protein ligand–substrate interactions in outer-membrane proteins with DEER/PELDOR spectroscopy
Nature Protocols (2019)
-
Paramagnetic-iterative relaxation matrix approach: extracting PRE-restraints from NOESY spectra for 3D structure elucidation of biomolecules
Journal of Biomolecular NMR (2019)
-
High-Frequency Electron Paramagnetic Resonance Spectroscopy of Nitroxide-Functionalized Nanodiamonds in Aqueous Solution
Cell Biochemistry and Biophysics (2017)
-
Site-directed spin-labeling of nucleotides and the use of in-cell EPR to determine long-range distances in a biologically relevant environment
Nature Protocols (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.