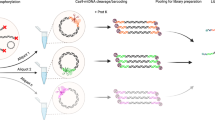
Abstract
Organelle genomics has become an increasingly important research field, with applications in molecular modeling, phylogeny, taxonomy, population genetics and biodiversity. Typically, research projects involve the determination and comparative analysis of complete mitochondrial and plastid genome sequences, either from closely related species or from a taxonomically broad range of organisms. Here, we describe two alternative organelle genome sequencing protocols. The “random genome sequencing” protocol is suited for the large majority of organelle genomes irrespective of their size. It involves DNA fragmentation by shearing (nebulization) and blunt-end cloning of the resulting fragments into pUC or BlueScript-type vectors. This protocol excels in randomness of clone libraries as well as in time and cost-effectiveness. The “long-PCR-based genome sequencing” protocol is specifically adapted for DNAs of low purity and quantity, and is particularly effective for small organelle genomes. Library construction by either protocol can be completed within 1 week.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Boer, P.H. & Gray, M.W. Scrambled ribosomal RNA gene pieces in Chlamydomonas reinhardtii mitochondrial DNA. Cell 55, 399–411 (1988).
Burger, G., Forget, L., Zhu, Y., Gray, M.W. & Lang, B.F. Unique mitochondrial genome architecture in unicellular relatives of animals. Proc. Natl. Acad. Sci. USA 100, 892–897 (2003).
Marande, W., Lukes, J. & Burger, G. Unique mitochondrial genome structure in diplonemids, the sister group of kinetoplastids. Eukaryot. Cell 4, 1137–1146 (2005).
Gray, M.W. Diversity and evolution of mitochondrial RNA editing systems. IUBMB Life 55, 227–233 (2003).
Gray, M.W. & Covello, P.S. RNA editing in plant mitochondria and chloroplasts. FASEB J. 7, 64–71 (1993).
Michel, F., Jacquier, A. & Dujon, B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie 64, 867–881 (1982).
Dujon, B., Colleaux, L., Jacquier, A., Michel, F. & Monteilhet, C. Mitochondrial introns as mobile genetic elements: the role of intron-encoded proteins. Basic Life Sci. 40, 5–27 (1986).
Lang, B.F., Gray, M.W. & Burger, G. Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 33, 351–397 (1999).
Gray, M.W., Lang, B.F. & Burger, G. Mitochondria of protists. Annu. Rev. Genet. 38, 477–524 (2004).
Bonen, L. & Vogel, J. The ins and outs of group II introns. Trends Genet. 17, 322–331 (2001).
Burger, G., Gray, M.W. & Lang, B.F. Mitochondrial genomes—anything goes. Trends Genet. 19, 709–716 (2003).
Bankier, A.T., Weston, K.M. & Barrell, B.G. Random cloning and sequencing by the M13/dideoxynucleotide chain termination method. Methods Enzymol. 155, 51–93 (1987).
Mullis, K.B. & Faloona, F.A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 155, 335–350 (1987).
Okpodu, C.M., Robertson, D., Boss, W.F., Togasaki, R.K. & Surzycki, S.J. Rapid isolation of nuclei from carrot suspension culture cells using a BioNebulizer. Biotechniques 16, 154–159 (1994).
Surzycki, S. in The International Conference on the Status and Future of Research on the Human Genome. Human Genome II (San Diego, CA), pp.51 (1990).
Ewing, B. & Green, P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8, 186–194 (1998).
Roe, B., Crabtree, J.S. & Khan, A.S. DNA Isolation and Sequencing (John Wiley and Sons, New York, 1996).
Cheng, S., Fockler, C., Barnes, W.M. & Higuchi, R. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc. Natl. Acad. Sci. USA 91, 5695–5699 (1994).
Boore, J.L., Macey, J.R. & Medina, M. Sequencing and comparing whole mitochondrial genomes of animals. Methods Enzymol. 395, 311–348 (2005).
Jansen, R.K. et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 395, 348–384 (2005).
Saghai-Maroof, M.A., Soliman, K.M., Jorgensen, R.A. & Allard, R.W. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 81, 8014–8018 (1984).
Berntson, E.A., France, S.C. & Mullineaux, L.S. Phylogenetic relationships within the class Anthozoa (phylum Cnidaria) based on nuclear 18S rDNA sequences. Mol. Phylogenet. Evol. 13, 417–433 (1999).
Palumbi,, S.R. in Molecular Phylogenetics (eds. Hillis, D.M., Moritz, C. & Mable, B.K.) 205–247 (Sinauer Associates Inc., Sunderland, MA, 1996).
Rozen, S. & Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386 (2000).
Robertson, J.M. & Walsh-Weller, J. An introduction to PCR primer design and optimization of amplification reactions. Methods Mol. Biol. 98, 121–154 (1998).
Lavrov, D.V., Forget, L., Kelly, M. & Lang, B.F. Mitochondrial genomes of two demosponges provide insights into an early stage of animal evolution. Mol. Biol. Evol. 22, 1231–1239 (2005).
Wesley, U.V. & Wesley, C.S. Rapid directional walk within DNA clones by step-out PCR. Methods Mol. Biol. 67, 279–285 (1997).
Shao, Z., Graf, S., Chaga, O.Y. & Lavrov, D.V. Mitochondrial genome of the moon jelly Aurelia aurita (Cnidaria, Scyphozoa): a linear DNA molecule encoding a putative DNA-dependent DNA polymerase. Gene 381, 92–101 (2006).
Fire, A. & Xu, S.Q. Rolling replication of short DNA circles. Proc. Natl. Acad. Sci. USA 92, 4641–4645 (1995).
Sambrook, J., Fritsch, E.F. & Maniatis, T. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, New York, NY, 1989).
Inoue, H., Nojima, H. & Okayama, H. High efficiency transformation of Escherichia coli with plasmids. Gene 96, 23–28 (1990).
Kocher, T.D. et al. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. USA 86, 6196–6200 (1989).
Boore, J.L. & Brown, W.M. Mitochondrial genomes of Galathealinum, Helobdella, and Platynereis: sequence and gene arrangement comparisons indicate that Pogonophora is not a phylum and Annelida and Arthropoda are not sister taxa. Mol. Biol. Evol. 17, 87–106 (2000).
Folmer, O., Black, M., Hoeh, W., Lutz, R. & Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299 (1994).
Lavrov, D.V., Brown, W.M. & Boore, J.L. Phylogenetic position of the Pentastomida and (pan)crustacean relationships. Proc. Biol. Sci. 271, 537–544 (2004).
Don, R.H., Cox, P.T., Wainwright, B.J., Baker, K. & Mattick, J.S. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19, 4008 (1991).
Kusukawa, N., Uemori, T., Asada, K. & Kato, I. Rapid and reliable protocol for direct sequencing of material amplified by the polymerase chain reaction. Biotechniques 9, 66–68 70, 72 (1990).
Acknowledgements
We thank Shona Teijeiro for critical reading of the manuscript and for help in the preparation of the figures. This work was supported by the Canadian Institute of Health Research (B.F.L., grant MOP-42475; G.B., grant MOP-79309; postdoctoral fellowship to D.V.L.). Interaction support from the Canadian Institute of Advanced Research, Program in Evolutionary Biology (G.B. and B.F.L.) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Burger, G., Lavrov, D., Forget, L. et al. Sequencing complete mitochondrial and plastid genomes. Nat Protoc 2, 603–614 (2007). https://doi.org/10.1038/nprot.2007.59
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.59
This article is cited by
-
Microfluidic continuous flow DNA fragmentation based on a vibrating sharp-tip
Microfluidics and Nanofluidics (2022)
-
The complete mitochondrial genome of a parasite at the animal-fungal boundary
Parasites & Vectors (2020)
-
An improved method for chloroplast genome sequencing in non-model forest tree species
Tree Genetics & Genomes (2015)
-
Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species
Scientific Reports (2015)
-
Mitochondrial genomes of the Baltic clam Macoma balthica(Bivalvia: Tellinidae): setting the stage for studying mito-nuclear incompatibilities
BMC Evolutionary Biology (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.