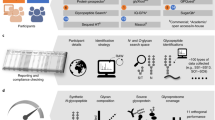
Abstract
Discovery of biomarker patterns using proteomic techniques requires examination of large numbers of patient and control samples, followed by data mining of the molecular read-outs (e.g., mass spectra). Adequate signal processing and statistical analysis are critical for successful extraction of markers from these data sets. The protocol, specifically designed for use in conjunction with MALDI-TOF-MS-based serum peptide profiling, is a data analysis pipeline, starting with transfer of raw spectra that are interpreted using signal processing algorithms to define suitable features (i.e., peptides). We describe an algorithm for minimal entropy-based peak alignment across samples. Peak lists obtained in this way, and containing all samples, all peptide features and their normalized MS-ion intensities, can be evaluated, and results validated, using common statistical methods. We recommend visual inspection of the spectra to confirm all results, and have written freely available software for viewing and color-coding of spectral overlays.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout












Similar content being viewed by others
References
Qian, W.J., Jacobs, J.M., Liu, T., Camp, D.G. & Smith, R.D., 2nd . Advances and challenges in liquid chromatography–mass spectrometry-based proteomics profiling for clinical applications. Mol. Cell Proteomics 5, 1727–1744 (2006).
Villanueva, J. et al. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J. Clin. Invest. 116, 271–284 (2006).
Villanueva, J. et al. Serum peptidome patterns that distinguish metastatic thyroid carcinoma from cancer-free controls are unbiased by gender and age. Mol. Cell Proteomics 5, 1840–1852 (2006).
Issaq, H.J., Conrads, T.P., Prieto, D.A., Tirumalai, R. & Veenstra, T.D. SELDI-TOF MS for diagnostic proteomics. Anal. Chem. 75, 148A–155A (2003).
Petricoin, E.F. & Liotta, L.A. SELDI-TOF-based serum proteomic pattern diagnostics for early detection of cancer. Curr. Opin. Biotechnol. 15, 24–30 (2004).
Koomen, J.M. et al. Direct tandem mass spectrometry reveals limitations in protein profiling experiments for plasma biomarker discovery. J. Proteome Res. 4, 972–981 (2005).
Richter, R. et al. Composition of the peptide fraction in human blood plasma: database of circulating human peptides. J. Chromatogr. B 726, 25–35 (1999).
Gao, J., Opiteck, G.J., Friedrichs, M.S., Dongre, A.R. & Hefta, S.A. Changes in the protein expression of yeast as a function of carbon source. J. Proteome Res. 2, 643–649 (2003).
Fach, E.M. et al. In vitro biomarker discovery for atherosclerosis by proteomics. Mol. Cell Proteomics 3, 1200–1210 (2004).
Wang, W. et al. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal. Chem. 75, 4818–4826 (2003).
Li, X.J. et al. A tool to visualize and evaluate data obtained by liquid chromatography-electrospray ionization-mass spectrometry. Anal. Chem. 76, 3856–3860 (2004).
Chen, S.S. et al. Improving mass and liquid chromatography based identification of proteins using bayesian scoring. J. Proteome Res. 4, 2174–2184 (2005).
Silva, J.C. et al. Quantitative proteomic analysis by accurate mass retention time pairs. Anal. Chem. 77, 2187–2200 (2005).
Jaitly, N. et al. Robust algorithm for alignment of liquid chromatography–mass spectrometry analyses in an accurate mass and time tag data analysis pipeline. Anal. Chem. 78, 7397–7409 (2006).
Wang, P. et al. A statistical method for chromatographic alignment of LC–MS data. Biostatistics (2006).
Gillette, M.A., Mani, D.R. & Carr, S.A. Place of pattern in proteomic biomarker discovery. J. Proteome Res. 4, 1143–1154 (2005).
Adam, B.L. et al. Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res. 62, 3609–3614 (2002).
Yanagisawa, K. et al. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet 362, 433–439 (2003).
Tibshirani, R. et al. Sample classification from protein mass spectrometry, by “peak probability contrasts”. Bioinformatics 20, 3034–3044 (2004).
Villanueva, J. et al. Serum peptide profiling by magnetic particle-assisted, automated sample processing and MALDI-TOF mass spectrometry. Anal. Chem. 76, 1560–1570 (2004).
Villanueva, J., Lawlor, K., Toledo-Crow, R. & Tempst, P. Automated serum peptide profiling. Nat. Prot. 1, 880–891 (2006).
DeNoyer, L. & Dodd, J. Smoothing and Derivatives in Spectroscopy. Vol. 3 (John Wiley and Sons, Chichester, UK, 2002).
Bylund, D., Danielsson, R., Malmquist, G. & Markides, K.E. Chromatographic alignment by warping and dynamic programming as a pre-processing tool for PARAFAC modelling of liquid chromatography–mass spectrometry data. J. Chromatogr. A 961, 237–244 (2002).
Villanueva, J. et al. Correcting common errors in identifying cancer-specific serum peptide signatures. J. Proteome Res. 4, 1060–1072 (2005).
Shannon, C.E. A mathematical theory of communication. Bell System Tech. J. 27, 379–423 (1948).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Method 1
Mass Spectra Analysis (PDF 447 kb)
Supplementary Method 2
Customized macro to convert Bruker files to ascii files (ZIP 2 kb)
Rights and permissions
About this article
Cite this article
Villanueva, J., Philip, J., DeNoyer, L. et al. Data analysis of assorted serum peptidome profiles. Nat Protoc 2, 588–602 (2007). https://doi.org/10.1038/nprot.2007.57
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.57
This article is cited by
-
A survey on cancer prediction and detection with data analysis
Innovations in Systems and Software Engineering (2020)
-
Detection of oesophageal cancer biomarkers by plasma proteomic profiling of human cell line xenografts in response to chemotherapy
British Journal of Cancer (2010)
-
A strategy with label-free quantification of the targeted peptides for quantitative peptidome analysis of human serum
Science China Chemistry (2010)
-
Precipitation and selective extraction of human serum endogenous peptides with analysis by quadrupole time-of-flight mass spectrometry reveals posttranslational modifications and low-abundance peptides
Analytical and Bioanalytical Chemistry (2010)
-
Prediction of outcome of non-small cell lung cancer patients treated with chemotherapy and bortezomib by time-course MALDI-TOF-MS serum peptide profiling
Proteome Science (2009)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.