Abstract
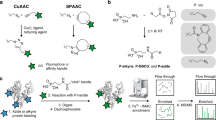
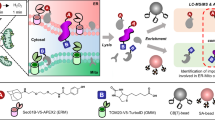
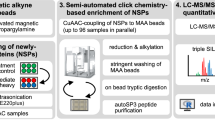
A major aim of proteomics is the identification of proteins in a given proteome at a given metabolic state. This protocol describes the step-by-step labeling, purification and detection of newly synthesized proteins in mammalian cells using the non-canonical amino acid azidohomoalanine (AHA). In this method, metabolic labeling of newly synthesized proteins with AHA endows them with the unique chemical functionality of the azide group. In the subsequent click chemistry tagging reaction, azide-labeled proteins are covalently coupled to an alkyne-bearing affinity tag. After avidin-based affinity purification and on-resin trypsinization, the resulting peptide mixture is subjected to tandem mass spectrometry for identification. In combination with deuterated leucine-based metabolic colabeling, candidate proteins can be immediately validated. Bioorthogonal non-canonical amino-acid tagging can be combined with any subcellular fractionation, immunopurification or other proteomic method to identify specific subproteomes, thereby reducing sample complexity and enabling the identification of subtle changes in a proteome. This protocol can be completed in 5 days.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pandey, A. & Mann, M. Proteomics to study genes and genomes. Nature 405, 837–846 (2000).
de Godoy, L. et al. Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol. 7, R50 (2006).
Mootha, V.K. et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115, 629–640 (2003).
Andersen, J.S. et al. Nucleolar proteome dynamics. Nature 433, 77–83 (2005).
Collins, M.O. et al. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J. Neurochem. 97 (suppl. 1): 16–23 (2006).
Schrimpf, S.P. et al. Proteomic analysis of synaptosomes using isotope-coded affinity tags and mass spectrometry. Proteomics 5, 2531–2541 (2005).
Gavin, A.C. et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 (2002).
Ho, Y. et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 (2002).
Chang, E.J., Archambault, V., McLachlin, D.T., Krutchinsky, A.N. & Chait, B.T. Analysis of protein phosphorylation by hypothesis-driven multiple-stage mass spectrometry. Anal. Chem. 76, 4472–4483 (2004).
Garcia, B.A., Shabanowitz, J. & Hunt, D.F. Analysis of protein phosphorylation by mass spectrometry. Methods 35, 256–264 (2005).
Peters, E.C., Brock, A. & Ficarro, S.B. Exploring the phosphoproteome with mass spectrometry. Mini Rev. Med. Chem. 4, 313–324 (2004).
Mann, M. & Jensen, O.N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21, 255–261 (2003).
Xu, P. & Peng, J. Dissecting the ubiquitin pathway by mass spectrometry. Biochim. Biophys. Acta 1764, 1940–1947 (2006).
Freeman, W.M. & Hemby, S.E. Proteomics for protein expression profiling in neuroscience. Neurochem. Res. 29, 1065–1081 (2004).
Lilley, K.S. & Friedman, D.B. All about DIGE: quantification technology for differential-display 2D-gel proteomics. Expert Rev. Proteomics 1, 401–409 (2004).
Gygi, S.P. et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999 (1999).
Ross, P.L. et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 (2004).
Washburn, M.P., Ulaszek, R., Deciu, C., Schieltz, D.M. & Yates, J.R., 3rd . Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Anal. Chem. 74, 1650–1657 (2002).
Ong, S.E. et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 (2002).
Dieterich, D.C., Link, A.J., Graumann, J., Tirrell, D.A. & Schuman, E.M. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. USA 103, 9482–9487 (2006).
Prescher, J.A. & Bertozzi, C.R. Chemistry in living systems. Nat. Chem. Biol. 1, 13–21 (2005).
Griffin, R.J. The medicinal chemistry of the azido group. Prog. Med. Chem. 31, 121–232 (1994).
Kho, Y. et al. A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc. Natl. Acad. Sci. USA 101, 12479–12484 (2004).
Kiick, K.L., Saxon, E., Tirrell, D.A. & Bertozzi, C.R. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc. Natl. Acad. Sci. USA 99, 19–24 (2002).
Link, A.J. & Tirrell, D.A. Cell surface labeling of Escherichia coli via copper(I)-catalyzed [3+2] cycloaddition. J. Am. Chem. Soc. 125, 11164–11165 (2003).
Link, A.J., Vink, M.K. & Tirrell, D.A. Presentation and detection of azide functionality in bacterial cell surface proteins. J. Am. Chem. Soc. 126, 10598–10602 (2004).
Luchansky, S.J., Goon, S. & Bertozzi, C.R. Expanding the diversity of unnatural cell-surface sialic acids. Chembiochem 5, 371–374 (2004).
Luchansky, S.J. et al. Constructing azide-labeled cell surfaces using polysaccharide biosynthetic pathways. Methods Enzymol. 362, 249–272 (2003).
Mahal, L.K., Yarema, K.J. & Bertozzi, C.R. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science 276, 1125–1128 (1997).
Saxon, E. et al. Investigating cellular metabolism of synthetic azidosugars with the Staudinger ligation. J. Am. Chem. Soc. 124, 14893–14902 (2002).
Rostovtsev, V.V., Green, L.G., Fokin, V.V. & Sharpless, K.B. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem. Int. Ed. Engl. 41, 2596–2599 (2002).
Mangold, J.B., Mischke, M.R. & LaVelle, J.M. Azidoalanine mutagenicity in Salmonella: effect of homologation and alpha-methyl substitution. Mutat. Res. 216, 27–33 (1989).
Wang, Q. et al. Bioconjugation by copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 125, 3192–3193 (2003).
Eng, J., McCormack, A.L. & Yates, A.J. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Perkins, D.N., Pappin, D.J., Creasy, D.M. & Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999).
Craig, R. & Beavis, R.C. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 (2004).
Fenyo, D. & Beavis, R.C. A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Anal. Chem. 75, 768–774 (2003).
Tabb, D.L., McDonald, W.H. & Yates, J.R., 3rd . DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 (2002).
Keller, A., Nesvizhskii, A.I., Kolker, E. & Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 (2002).
Nesvizhskii, A.I., Keller, A., Kolker, E. & Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 (2003).
Link, A.J. et al. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17, 676–682 (1999).
Washburn, M.P., Wolters, D. & Yates, J.R., 3rd . Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 (2001).
Schieltz, D.M. & Washburn, M.P. Analysis of complex protein mixtures using multidimensional protein identification technology (MuDPIT). Cold Spring Harbor Protocols 2006 (10.1101/pdb.prot4555).
Acknowledgements
We thank S.A. Kim and E.H. Friedrich for critical reading of the manuscript. This work was supported by the Howard Hughes Medical Institute, the Beckman Institute at the California Institute of Technology and NIH (R21DA020589 to E.M.S.). MS analysis was performed at the MS facility of the laboratory of R.J. Deshaies (Howard Hughes Medical Institute, Caltech), which is supported by the Beckman Institute at Caltech and a grant from the Department of Energy to R.J.D. and Barbara J. Wold. D.C.D. is supported by the German Academy for Natural Scientists Leopoldina (BMBF-LPD9901/8-95). J.G. was supported by R.J. Deshaies through Howard Hughes Medical Institute funds. A.J.L. was supported by a National Science Foundation Graduate Research Fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Dieterich, D., Lee, J., Link, A. et al. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc 2, 532–540 (2007). https://doi.org/10.1038/nprot.2007.52
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.52
This article is cited by
-
Deoxynivalenol triggers the expression of IL-8-related signaling cascades and decreases protein biosynthesis in primary monocyte-derived cells
Mycotoxin Research (2024)
-
Modulation of cellular transcriptome and proteome composition by azidohomoalanine—implications on click chemistry–based secretome analysis
Journal of Molecular Medicine (2023)
-
Effects of Pregnancy-Specific Glycoproteins on Trophoblast Motility in Three-Dimensional Gelatin Hydrogels
Cellular and Molecular Bioengineering (2022)
-
Extracellular Matrix by Design: Native Biomaterial Fabrication and Functionalization to Boost Tissue Regeneration
Regenerative Engineering and Translational Medicine (2022)
-
Comprehensive and systematic characterization of multi-functionalized cisplatin nano-conjugate: from the chemistry and proteomic biocompatibility to the animal model
Journal of Nanobiotechnology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.