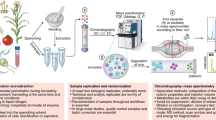
Abstract
Flow injection electrospray–mass spectrometry (FIE–MS) is finding utility as a first-pass metabolite fingerprinting tool in many research fields. We provide a protocol that has proved reliable in large-scale research projects involving diverse sample matrices originating from plants, microbes and mammalian biofluids. Using Brachypodium leaf material as an example matrix all steps are summarized from sample extraction to data quality assessment. Alternative procedures for dealing with other common matrices such as bloods and urine are included. With little sample pretreatment, no chromatography and instrument cycle times of <5 min it is feasible to analyze >1,000 samples per week. Analysis of a typical batch of 240 samples (including first-pass data analysis) can be accomplished within 48 h.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout











Similar content being viewed by others
References
Martin, D.B. & Nelson, P.S. From genomics to proteomics: techniques and applications in cancer research. Trends Cell Biol. 11, S60–S65 (2001).
Saghatelian, A. & Cravatt, B.F. Global strategies to integrate the proteome and metabolome. Curr. Opin. Chem. Biol. 9, 62–68 (2005).
Murray, D.B., Beckmann, M. & Kitano, H. Regulation of yeast oscillatory dynamics. Proc. Natl. Acad. Sci. USA 104, 2241–2246 (2007).
Catchpole, G.S. et al. Hierarchical metabolomics demonstrates substantial compositional similarity between genetically modified and conventional potato crops. Proc. Natl. Acad. Sci. USA 102, 14458–14462 (2005).
Bino, R.J. et al. Potential of metabolomics as a functional genomics tool. Trends Plant Sci. 9, 418–425 (2004).
Fiehn, O. Metabolomics–the link between genotypes and phenotypes. Plant Mol. Biol. 48, 155–171 (2002).
Lisec, J., Schauer, N., Kopka, J., Willmitzer, L. & Fernie, A.R. Gas chromatography mass spectrometry–based metabolite profiling in plants. Nat. Protoc. 1, 387–396 (2006).
Kaderbhai, N.N., Broadhurst, D.I., Ellis, D.I., Goodacre, R. & Kell, D.B. Functional genomics via metabolic footprinting: monitoring metabolite secretion by Escherichia coli tryptophan metabolism mutants using FT-IR and direct injection electrospray mass spectrometry. Comp. Funct. Genomics 4, 376–391 (2003).
Enot, D.P., Beckmann, M., Overy, D. & Draper, J. Predicting interpretability of metabolome models based on behavior, putative identity, and biological relevance of explanatory signals. Proc. Natl. Acad. Sci. USA 103, 14865–14870 (2006).
Ward, J.L., Harris, C., Lewis, J. & Beale, M.H. Assessment of H-1 NMR spectroscopy and multivariate analysis as a technique for metabolite fingerprinting of Arabidopsis thaliana. Phytochemistry 62, 949–957 (2003).
Roessner, U., Wagner, C., Kopka, J., Trethewey, R.N. & Willmitzer, L. Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 23, 131–142 (2000).
Taylor, J., King, R.D., Altmann, T. & Fiehn, O. Application of metabolomics to plant genotype discrimination using statistics and machine learning. Bioinformatics 18, S241–S248 (2002).
Wagner, C., Sefkow, M. & Kopka, J. Construction and application of a mass spectral and retention time index database generated from plant GC/EI-TOF-MS metabolite profiles. Phytochemistry 62, 887–900 (2003).
Tolstikov, V.V., Lommen, A., Nakanishi, K., Tanaka, N. & Fiehn, O. Monolithic silica-based capillary reversed-phase liquid chromatography/electrospray mass spectrometry for plant metabolomics. Anal. Chem. 75, 6737–6740 (2003).
Tolstikov, V.V. & Fiehn, O. Analysis of highly polar compounds of plant origin: combination of hydrophilic interaction chromatography and electrospray ion trap mass spectrometry. Anal. Biochem. 301, 298–307 (2002).
Goodacre, R., Vaidyanathan, S., Dunn, W.B., Harrigan, G.G. & Kell, D.B. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 22, 245–252 (2004).
Sumner, L.W., Mendes, P. & Dixon, R.A. Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry 62, 817–836 (2003).
Fiehn, O. et al. Metabolite profiling for plant functional genomics. Nat. Biotechnol. 18, 1157–1161 (2000).
Dear, G.J., James, A.D. & Sarda, S. Ultra-performance liquid chromatography coupled to linear ion trap mass spectrometry for the identification of drug metabolites in biological samples. Rapid Commun. Mass Spectrom. 20, 1351–1360 (2006).
Startin, J.R., Hird, S.J. & Sykes, M.D. Determination of ethylenethiourea (ETU) and propylenethiourea (PTU) in foods by high performance liquid chromatography-atmospheric pressure chemical ionisation-medium-resolution mass spectrometry. Food Addit. Contam. 22, 245–250 (2005).
Wang, W. et al. Quantification of proteins and metabolites by mass spectrometry without isotopic labelling or spiked standards. Anal. Chem. 75, 4818–4826 (2003).
Smedsgaard, J., Hansen, M.E. & Frisvad, J.C. Classification of terverticillate Penicillia by electrospray mass spectrometric profiling. Stud. Mycol. 49, 243–251 (2004).
Goodacre, R. et al. Detection of the dipicolinic acid biomarker in Bacillus spores using Curie-point pyrolysis mass spectrometry and fourier transform infrared spectroscopy. Anal. Chem. 72, 119–127 (2000).
Nicholson, J.K. & Wilson, I.D. Opinion: understanding 'global' systems biology: metabonomics and the continuum of metabolism. Nat. Rev. Drug Discov. 2, 668–676 (2003).
Allen, J. et al. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat. Biotechnol. 21, 692–696 (2003).
Scholz, M., Gatzek, S., Sterling, A., Fiehn, O. & Selbig, J. Metabolite fingerprinting: detecting biological features by independent component analysis. Bioinformatics 20, 2447–2454 (2004).
Aharoni, A. et al. Nontargeted metabolome analysis by use of Fourier transform ion cyclotron mass spectrometry. OMICS 6, 217–234 (2002).
Smedsgaard, J. & Frisvad, J.C. Using direct electrospray mass spectrometry in taxonomy and secondary metabolite profiling of crude fungal extracts. J. Microbiol. Methods 25, 5–17 (1996).
Hopfgartner, G. et al. Triple quadrupole linear ion trap mass spectrometer for the analysis of small molecules and macromolecules. J. Mass Spectrom. 39, 845–855 (2004).
Zamfir, A.D. et al. Thin chip microsprayer system coupled to quadrupole time-of-flight mass spectrometer for glycoconjugate analysis. Lab Chip 5, 298–307 (2005).
Nielsen, K.F. & Smedsgaard, J. Fungal metabolite screening: database of 474 mycotoxins and fungal metabolites for dereplication by standardised liquid chromatography-UV-mass spectrometry methodology. J. Chromatogr. A 1002, 111–136 (2003).
Gorlach, E. & Richmond, R. Discovery of quasi-molecular ions in electrospray spectra by automated searching for simultaneous adduct mass differences. Anal. Chem. 71, 5557–5562 (1999).
Overy, D.P. et al. Explanatory signal interpretation and metabolite identification strategies for nominal mass FIE-MS metabolite fingerprints. Nat. Protoc. 3, 471–485 (2008).
Antignac, J.P. et al. The ion suppression phenomenon in liquid chromatography-mass spectrometry and its consequences in the field of residue analysis. Anal. Chim. Acta 529, 129–136 (2005).
van Hout, M.W., Niederländer, H.A., de Zeeuw, R.A. & de Jong, G.J. Ion suppression in the determination of clenbuterol in urine by solid-phase extraction atmospheric pressure chemical ionisation ion-trap mass spectrometry. Rapid Commun. Mass Spectrom. 17, 245–250 (2003).
Müller, C., Schäfer, P., Störtzel, M., Vogt, S. & Weinmann, W. Ion suppression effects in liquid chromatography-electrospray-ionisation transport-region collision induced dissociation mass spectrometry with different serum extraction methods for systematic toxicological analysis with mass spectra libraries. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 773, 47–52 (2002).
Annesley, T.M. Ion suppression in mass spectrometry. Clin. Chem. 49, 1041–1044 (2003).
Vaidyanathan, S., Kell, D.B. & Goodacre, R. Flow-injection electrospray ionization mass spectrometry of crude cell extracts for high-throughput bacterial identification. J. Am. Soc. Mass Spectrom. 13, 118–128 (2002).
Mazzella, N. et al. Use of electrospray ionization mass spectrometry for profiling of crude oil effects on the phospholipid molecular species of two marine bacteria. Rapid Commun. Mass Spectrom. 19, 3579–3588 (2005).
Favretto, D., Piovan, A., Filippini, R. & Caniato, R. Monitoring the production yields of vincristine and vinblastine in Catharanthus roseus from somatic embryogenesis. Semiquantitative determination by flow-injection electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 15, 364–369 (2001).
Zahn, J.A., Higgs, R.E. & Hilton, M.D. Use of direct-infusion electrospray mass spectrometry to guide empirical development of improved conditions for expression of secondary metabolites from actinomycetes. Appl. Environ. Microbiol. 67, 377–386 (2001).
Goodacre, R., York, E.V., Heald, J.K. & Scott, I.M. Chemometric discrimination of unfractionated plant extracts analyzed by electrospray mass spectrometry. Phytochemistry 62, 859–863 (2003).
Overy, S.A. et al. Application of metabolite profiling to the identification of traits in a population of tomato introgression lines. J. Exp. Bot. 56, 287–296 (2005).
Koulman, A. et al. High-throughput direct-infusion ion trap mass spectrometry: a new method for metabolomics. Rapid Commun. Mass Spectrom. 21, 421–428 (2007).
Rashed, M.S., Al-Ahaidib, L.Y., Aboul-Enein, H.Y., Al-Amoudi, M. & Jacob, M. Determination of L-pipecolic acid in plasma using chiral liquid chromatography-electrospray tandem mass spectrometry. Clin. Chem. 47, 2124–2130 (2001).
Kell, D.B., Darby, R.M. & Draper, J. Genomic computing. Explanatory analysis of plant expression profiling data using machine learning. Plant Physiol. 126, 943–951 (2001).
Kay, C.D., Mazza, G., Holub, B.J. & Wang, J. Anthocyanin metabolites in human urine and serum. Br. J. Nutr. 91, 933–942 (2004).
Beckmann, M. & Enot, D.P. Overy, D & Draper, J. Representation, comparison and interpretation of metabolome fingerprint data for total composition analysis and quality trait investigation in potato plants. J. Agric. Food. Chem. 55, 3444–3451 (2007).
Parker, D. et al. Rice blast infection of Brachypodium distachyon as a model system to study dynamic host/pathogen interactions. Nat. Protoc. 3, 435–445 (2008).
Enot, D. et al. Preprocessing, classification modeling and feature selection using flow injection electrospray mass spectrometry metabolite fingerprint data. Nat. Protoc. 3, 446–470 (2008).
Jenkins, H. et al. A proposed framework for the description of plant metabolomics experiments and their results. Nat. Biotechnol. 22, 1601–1606 (2004).
Castle, A.L., Fiehn, O., Kaddurah-Daouk, R. & Lindon, J.C. Metabolomics Standards Workshop and the development of international standards for reporting metabolomics experimental results. Brief. Bioinform. 7, 159–165 (2006).
Broadhurst, D.I. & Kell, D.B. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics 2, 171–196 (2006).
Bligh, E.G. & Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959).
Enot, D.P., Beckmann, M. & Draper, J. Detecting a difference–assessing generalisability when modelling metabolome fingerprint data in longer term studies of genetically modified plants. Metabolomics 3, 335–347 (2007).
Chernushevich, I.V., Loboda, A.V. & Thomson, B.A. An introduction to quadrupole-time-of-flight mass spectrometry. J. Mass Spectrom. 36, 849–865 (2001).
Nieuwoudt, H.H., Prior, B.A., Pretorius, I.S., Manley, M. & Bauer, F.F. Principal component analysis applied to Fourier transform infrared spectroscopy for the design of calibration sets for glycerol prediction models in wine and for the detection and classification of outlier samples. J. Agric. Food Chem. 52, 3726–3735 (2004).
Gullberg, J., Jonsson, P., Nordstrom, A., Sjostrom, M. & Moritz, T. Design of experiments: an efficient strategy to identify factors influencing extraction and derivatization of Arabidopsis thaliana samples in metabolomic studies with gas chromatography/mass spectrometry. Anal. Biochem. 331, 283–295 (2004).
R_Development_Core_Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, Vienna, 2006, http://www.R-project.org).
Acknowledgements
We thank all collaborators who provided valuable samples to develop this protocol and, in particular, Rob Darby for overarching support with laboratory infrastructure, equipment and materials. Metabolite analysis and statistical work (M.B. and D.E.) was partly funded by the Food Standards Agency (London) as part of its G02006 and G03012 projects and biological materials used in example data were generated as part of the UK Biotechnology and Biological Sciences Research council grant BBD0069531 (D.P. and E.D.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Beckmann, M., Parker, D., Enot, D. et al. High-throughput, nontargeted metabolite fingerprinting using nominal mass flow injection electrospray mass spectrometry. Nat Protoc 3, 486–504 (2008). https://doi.org/10.1038/nprot.2007.500
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.500
This article is cited by
-
Small molecule metabolites: discovery of biomarkers and therapeutic targets
Signal Transduction and Targeted Therapy (2023)
-
Metabolomic changes in polyunsaturated fatty acids and eicosanoids as diagnostic biomarkers in Mycobacterium avium ssp. paratuberculosis (MAP)-inoculated Holstein–Friesian heifers
Veterinary Research (2022)
-
Spectral binning as an approach to post-acquisition processing of high resolution FIE-MS metabolome fingerprinting data
Metabolomics (2022)
-
Metabolomic changes in Mycobacterium avium subsp. paratuberculosis (MAP) challenged Holstein–Friesian cattle highlight the role of serum amino acids as indicators of immune system activation
Metabolomics (2022)
-
Fast and sensitive flow-injection mass spectrometry metabolomics by analyzing sample-specific ion distributions
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.