Abstract
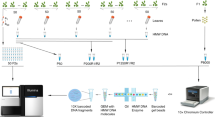
In most organisms, one crossover (CO) event inhibits the chances of another nearby event. The term used to describe this phenomenon is 'CO interference'. Here, we describe a protocol for quickly generating large data sets that are amenable to CO interference analysis in the flowering plant, Arabidopsis thaliana. We employ a visual assay that utilizes transgenic marker constructs encoding pollen-expressed fluorescent proteins of three colors in the quartet mutant background. In this genetic background, male meiotic products—the pollen grains—remain physically attached thereby facilitating tetrad analysis. We have developed a library of mapped marker insertions that, when crossed together, create adjacent intervals that can be rapidly and simultaneously screened for COs. This assay system is capable of detecting and differentiating single COs as well as two-, three- and four-strand double COs. We also describe how to analyze the data that are produced by this method. To generate and score a double interval in a wild-type and mutant background using this protocol will take 22–27 weeks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Zickler, D. & Kleckner, N. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32, 619–697 (1998).
Page, S.L. & Hawley, R.S. Chromosome choreography: the meiotic ballet. Science 301, 785–789 (2003).
Felsenstein, J. The evolutionary advantage of recombination. Genetics 78, 737–756 (1974).
Jones, G.H. & Franklin, F.C. Meiotic crossing-over: obligation and interference. Cell 126, 246–248 (2006).
Borner, G.V., Kleckner, N. & Hunter, N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117, 29–45 (2004).
Foss, E., Lande, R., Stahl, F.W. & Steinberg, C.M. Chiasma interference as a function of genetic distance. Genetics 133, 681–691 (1993).
Sturtevant, A.H. The behavior of chromosomes as studied through linkage. Z. Induct. Abstammungs-Vererbungsl. 13, 234–287 (1915).
Muller, H.J. The mechanism of crossing over. Am. Nat. 50, 193–434 (1916).
Hillers, K.J. Crossover interference. Curr. Biol. 14, R1036–R1037 (2004).
Copenhaver, G.P., Housworth, E.A. & Stahl, F.W. Crossover interference in Arabidopsis. Genetics 160, 1631–1639 (2002).
Housworth, E.A. & Stahl, F.W. Crossover interference in humans. Am. J. Hum. Genet. 73, 188–197 (2003).
Foss, E.J. & Stahl, F.W. A test of a counting model for chiasma interference. Genetics 139, 1201–1209 (1995).
McPeek, M.S. & Speed, T.P. Modeling interference in genetic recombination. Genetics 139, 1031–1044 (1995).
Drouaud, J. et al. Sex-specific crossover distributions and variations in interference level along Arabidopsis thaliana chromosome 4. PloS Genet. 3, e106 (2007).
Hawley, R.S., McKim, K.S. & Arbel, T. Meiotic segregation in Drosophila melanogaster females: molecules, mechanisms, and myths. Annu. Rev. Genet. 27, 281–317 (1993).
Francis, K.E. et al. A pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc. Natl. Acad. Sci. USA 104, 3913–3918 (2007).
Berchowitz, L.E., Francis, K.E., Bey, A.L. & Copenhaver, G.P. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PloS Genet. 3, e132 (2007).
Lam, S.Y. et al. Crossover interference on nucleolus organizing region-bearing chromosomes in Arabidopsis. Genetics 170, 807–812 (2005).
Malkova, A. et al. Gene conversion and crossing over along the 405-kb left arm of Saccharomyces cerevisiae chromosome VII. Genetics 168, 49–63 (2004).
Mortimer, R.K. & Hawthorne, D.C. Genetic mapping in Saccharomyces IV. Mapping of temperature-sensitive genes and use of disomic strains in localizing genes. Genetics 74, 33–54 (1973).
Fogel, S. & Mortimer, R.K. Recombination in yeast. Annu. Rev. Genet. 5, 219–236 (1971).
Yauk, C.L., Bois, P.R. & Jeffreys, A.J. High-resolution sperm typing of meiotic recombination in the mouse MHC beta gene. EMBO J 22, 1389–1397.
Tiemann-Boege, I., Calabrese, P., Cochran, D.M., Sokol, R. & Arnheim, N. High-resolution recombination patterns in a region of human chromosome 21 measured by sperm typing. PloS Genet. 2, e70 (2006).
Perkins, D.D. The detection of linkage in tetrad analysis. Genetics 38, 187–197 (1953).
Papazian, H.P. The analysis of tetrad data. Genetics 37, 175–188 (1952).
Hawley, R.S. Meiosis in living color: fluorescence-based tetrad analysis in Arabidopsis. Proc. Natl. Acad. Sci. USA 104, 3673–3674 (2007).
Copenhaver, G.P. A compendium of plant species producing pollen tetrads. J. North Carolina Acad. Sci. 121, 17–35 (2005).
Copenhaver, G.P., Browne, W.E. & Preuss, D. Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proc. Natl. Acad. Sci. USA 95, 247–252 (1998).
Jeffreys, A.J. et al. Meiotic recombination hot spots and human DNA diversity. Phil. Trans. R. Soc. Lond. B. Biol. Sci. 359, 141–152 (2004).
Janssens, F.A. Spermatogénese dans les batraciens. V. La théorie de la chiasmatypie, nouvelle interpretation des cinéses de maturation. Cellule 25, 387–411 (1909).
Higgins, J.D., Armstrong, S.J., Franklin, F.C. & Jones, G.H. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes. Dev. 18, 2557–2570 (2004).
de Boer, E., Dietrich, A.J., Hoog, C., Stam, P. & Heyting, C. Meiotic interference among MLH1 foci requires neither an intact axial element structure nor full synapsis. J. Cell. Sci. 120, 731–736 (2007).
de Boer, E., Stam, P., Dietrich, A.J., Pastink, A. & Heyting, C. Two levels of interference in mouse meiotic recombination. Proc. Natl. Acad. Sci. USA 103, 9607–9612 (2006).
Jones, G.H. The control of chiasma distribution. In Controlling Events In Meiosis (eds. Evans, C., & Dickinson, H.) 293–320 (The Company of Biologists, Cambridge, UK, 1984).
Carpenter, A.T. Recombination nodules and synaptonemal complex in recombination-defective females of Drosophila melanogaster. Chromosoma 75, 259–292 (1979).
Sherman, J.D. & Stack, S.M. Two-dimensional spreads of synaptonemal complexes from solanaceous plants. VI. High-resolution recombination nodule map for tomato (Lycopersicon esculentum). Genetics 141, 683–708 (1995).
Carpenter, A.T. Synaptonemal complex and recombination nodules in wild-type Drosophila melanogaster females. Genetics 92, 511–541 (1979).
Carpenter, A.T. Electron microscopy of meiosis in Drosophila melanogaster females: II. The recombination nodule—a recombination-associated structure at pachytene? Proc. Natl. Acad. Sci. USA 72, 3186–3189 (1975).
Sanchez Moran, E., Armstrong, S.J., Santos, J.L., Franklin, F.C. & Jones, G.H. Chiasma formation in Arabidopsis thaliana accession Wassileskija and in two meiotic mutants. Chromosome Res. 9, 121–128 (2001).
Wijeratne, A.J., Chen, C., Zhang, W., Timofejeva, L. & Ma, H. The Arabidopsis thaliana PARTING DANCERS gene encoding a novel protein is required for normal meiotic homologous recombination. Mol. Biol. Cell. 17, 1331–1343 (2006).
Preuss, D., Rhee, S.Y. & Davis, R.W. Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264, 1458–1460 (1994).
Copenhaver, G.P., Keith, K.C. & Preuss, D. Tetrad analysis in higher plants. A budding technology. Plant. Physiol 124, 7–26 (2000).
Twell, D., Yamaguchi, J. & McCormick, S. Pollen-specific gene expression in transgenic plants: coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109, 705–713 (1990).
Perkins, D.D. Crossing-over and interference in a multiply marked chromosome arm of Neurospora. Genetics 47, 1253–1274 (1962).
Stahl, F.W. & Lande, R. Estimating interference and linkage map distance from two-factor tetrad data. Genetics 139, 1449–1454 (1995).
Mezard, C., Vignard, J., Drouaud, J. & Mercier, R. The road to crossovers: plants have their say. Trends Genet. 23, 91–99.
Perkins, D.D. Biochemical mutants in the smut fungus Ustilago maydis. Genetics 34, 607–626 (1949).
Stahl, F.W. et al. Does crossover interference count in Saccharomyces cerevisiae? Genetics 168, 35–48 (2004).
Weigel, D. & Glazebrook, J. Arabidopsis: A Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2002).
Acknowledgements
We thank Corbin Jones and Frank Stahl for critical reading of this manuscript. We also thank the NSF (MCB-0618691) and DOE (DE-FGO2-05ER15651) for financial support.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Fig. 1
Blank tetrad classification template (PDF 255 kb)
Rights and permissions
About this article
Cite this article
Berchowitz, L., Copenhaver, G. Fluorescent Arabidopsis tetrads: a visual assay for quickly developing large crossover and crossover interference data sets. Nat Protoc 3, 41–50 (2008). https://doi.org/10.1038/nprot.2007.491
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.491
This article is cited by
-
MSH2 stimulates interfering and inhibits non-interfering crossovers in response to genetic polymorphism
Nature Communications (2023)
-
Epigenetic regulation during meiosis and crossover
Physiology and Molecular Biology of Plants (2023)
-
The megabase-scale crossover landscape is largely independent of sequence divergence
Nature Communications (2022)
-
HIGH CROSSOVER RATE1 encodes PROTEIN PHOSPHATASE X1 and restricts meiotic crossovers in Arabidopsis
Nature Plants (2021)
-
High temperatures alter cross-over distribution and induce male meiotic restitution in Arabidopsis thaliana
Communications Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.