Abstract
The increasing multi-resistance of pathogenic bacteria requires the development of novel classes of antibiotics. Antimicrobial host defense peptides represent one promising class. Here we describe a protocol for screening large numbers of peptides against any microbe of interest. Peptides synthesized on a cellulose support by peptide array technology can be added to a microbe that expresses the luxCDABE (luciferase) gene cassette. Any substance that decreases the energy level within the microbe will cause a quantifiable decrease in light production. The potency of the compound, at different concentrations, is reflected by the rate of decrease in luminescence. In conjunction with peptide array technology, the screening assay is rapid and high throughput and demonstrates good correlation with conventional killing or minimal inhibitory concentration assays performed with the same peptides synthesized by standard solid-phase peptide synthesis. The protocol can be completed in 3 d.
Similar content being viewed by others
Introduction
Antimicrobial substances play a critical role in the treatment of infectious diseases in both humans and animals. The rapid increase in antibiotic resistance has resulted in pressure to find antibiotics with novel modes of action for treating infections caused by multi-drug-resistant bacteria1. Natural products as well as chemically synthesized compounds have been used to search for novel antibiotics. Recent candidates as novel antibiotics are cationic antimicrobial (host defense) peptides2, and one is already in clinical phase III trials. Here we report a time- and cost-efficient screening assay for short antimicrobial peptides suitable for bioluminescent and non-bioluminescent microbes. This assay can be adapted for use with other antimicrobials, although the concentration of the antimicrobial substance, contact time of microbe and antibiotic and buffer/medium conditions for the microbes may require optimization.
Until recently, peptides were usually obtained from biological sources. Selected natural peptides were taken, and amino acid substitutions based on the sequence information or on the 3D structure of the peptides were made. These peptide variants were then synthesized by conventional solid- or solution-phase peptide chemistry. This approach limited the exploration of peptide diversity since it is very expensive (costing $100 or more per peptide screened). Conversely, larger numbers of peptides have been explored using biological or chemical peptide libraries.
In general, biological libraries such as phage3, bacterial4, or ribosome5 display are tricky to handle. In addition, these approaches create fusion peptides rather than isolated molecules and use only the gene-encoded amino acids. One advantage of these libraries is that the peptides are synthesized biologically, and therefore manual synthesis and expensive chemicals are not required. Furthermore, rounds of enrichment permit the potential selection of strongly active peptides. This is offset by the problem that information on optimal amino acid substitutions is contextual and not systematic, and also only minor information is gained at positions that do not lead to variants with improved activities. In addition, multiple substitutions may occur, thus limiting the ability to obtain systematic information for each position to optimize peptides in a rational fashion, e.g., leading to moderately active peptides with minimal inhibitory concentrations (MICs) versus Escherichia coli of 500 μg ml−1 for linear 10-mer peptides from phage display3 or 25 μg ml−1 from ribosomal display5. (The MIC of an antibacterial or antifungal substance is defined as the maximum dilution of the substance that will still inhibit the growth of a test microorganism under defined test conditions.)
In contrast to biological peptide libraries, chemical peptide libraries are not restricted to gene-encoded amino acids; therefore, more than 600 commercially available building blocks (non-gene-coded amino acids, D-amino acids and non-amino acids) can be used. Screening large numbers of peptides using peptide synthesis on resin is labor intense and expensive. At only approximately 1% of the cost per peptide compared to the conventional peptide synthesis on resin, the synthesis of arrays of peptides on cellulose using SPOT technology represents a powerful tool for characterizing or screening large numbers of peptides for antimicrobial activity6,7,8,9. Using SPOT synthesis, up to 1,000 peptides per 20 × 29–cm2 cellulose sheet can be synthesized in a highly parallel and addressable manner. SPOT technology allows peptides to be synthesized up to a length of 50 aa, with an optimal range of 6–18 aa. Peptide arrays can be produced, given the appropriate instrumentation, by manual, semi-automated or fully automated syntheses. The instruments are commercially available (Intavis, Köln, Germany). The principal array strategies provided by peptide SPOT synthesis are peptide scan, substitution analyses, length analyses, random libraries and combinatorial libraries. For a review of these strategies see ref. 7. In addition, an approach to optimizing antimicrobial peptides using scrambled peptide sequences was described recently10. When synthesized via SPOT technology, peptides are covalently bound to cellulose via their carboxy termini through a linker. Either they can then be tested for activity after cleavage from the membrane (for cleavage treatments, see ref. 7) or, as we have recently learned, some peptides remain active even when still covalently attached to the cellulose surface (K. Hilpert, M. Elliott, S.W. Farmer, R. Volkmer-Engert, P. Henklein and R.E.W. Hancock, manuscript in preparation). The ability to access such techniques for optimization of antimicrobial peptides led to 8–12 aa antimicrobial peptides with MICs of 0.5–2 μg ml−1 against a broad spectrum of bacteria and of 3 μg ml−1 against the fungus Candida albicans. In addition, peptide libraries synthesized on pins were used to screen for antimicrobial activity11.
The key to testing peptides for their antimicrobial activity is to have a flexible and quantifiable method that permits rapid screening of large numbers of peptides synthesized on cellulose. Direct testing of killing activity is possible but cumbersome, and both the sensitivity and dynamic range can be limited. Therefore we adapted a luminescence inhibition assay that has been used previously for a variety of bacteria and antimicrobial agents12,13,14,15,16, can be performed in small volumes and is rapid (giving a readout of activity within 30 min or less), sensitive and quantitative8,15. This assay relies on the genes expressed from a luxCDABE cassette. The genes luxCDE encode a fatty acid reductase complex that results in the synthesis of a fatty aldehyde substrate for the luminescence reaction catalyzed by the luciferase subunits encoded by luxAB. To generate light, bacteria harboring this cassette, which can be introduced via a transposon or plasmid, must supply only reduced flavin mononucleotide (FMNH2) and molecular oxygen. In the case that the energy level of the microbe is disturbed, the resulting drop in FMNH2 and therefore the change in luminescence intensity can be monitored. The inhibition of luminescence, which can be sensitively measured using a luminometer in a multiplex (e.g., microtiter plate) format, provides a measurement of inhibition of microbial metabolism. In this protocol the Gram-negative bacterium Pseudomonas aeruginosa is used to screen for antimicrobial activity of peptides. In particular, we chose to use a P. aeruginosa isolate, strain H1001, with a Tn5-luxCDABE transposon inserted into the fliC gene, as this mutant grows normally and expresses luminescence in an abundant and constitutive fashion, and because Pseudomonas is one of the most antibiotic-resistant pathogens in our environment and thus an important target for antibiotic development. However, any bacterium transformed with a luxCDABE cassette17 could be used. In principle, the assay described here could be performed using microbes transformed with any other energy-dependent light-producing enzyme (such as firefly luciferase in combination with luciferin).
Experimental design
The limitations of reliability of SPOT technology have been described by several authors18,19,20. Since purity and quantity of the peptides synthesized on cellulose may vary, all results achieved using this method need to be confirmed with peptides produced by conventional solid-phase peptide synthesis on resin. It is worth noting that antibacterial activity assays with soluble peptides usually confirm the activity of peptides synthesized on cellulose8,10. However, the correct design and preparation of the assay are critical to the success of the method.
Despite attempts to control all parameters, including the use of a standardized growth procedure for the indicator bacteria, daily variations in bioluminescence and consequent alteration in the observed killing activity may occur. To compare data from one day to the next, it is important that the same negative and positive controls are added to each microtiter plate. These control peptides must be synthesized on the same membrane as the one used to produce all other peptides in a given screen. Consequently, two control peptides, one positive control and one negative control, must be synthesized for every ten peptides designated for screening (Table 1). Additional positive and negative control peptides, as well as some randomly selected peptides, all synthesized on the same cellulose membrane, can support the process of fine-tuning the assay for specific requirements. As a test of the procedure conditions for the individual peptide set, we recommend performing the whole protocol step by step with only the negative and positive control peptides as well as randomly selected peptides from the screening set first, to avoid wasting peptides designated for screening. These additional peptides can also be used for HPLC and mass spectroscopy analysis to assess the quality of the synthesis. To provide greater security regarding the antimicrobial assay and to estimate the concentration of the peptides synthesized on cellulose, we recommend occasionally adding a positive control (resin synthesized) at known concentrations to compare its activity with the activity of the same peptide synthesized on cellulose.
The method here assumes that peptide arrays have been prepared in the laboratory or purchased and that an appropriate cleavage step has been undertaken to sever the covalent linkage with the cellulose membrane, releasing the peptides. A complete protocol to synthesize peptides on cellulose membranes is described in ref. 21. There are two critical steps in the peptide synthesis protocol that may influence the screen for antimicrobial activity. First, it is important to mark the center of the synthesized peptide spot before the last step of peptide synthesis. This will improve accuracy in the subsequent punching out of the peptides from the cellulose membrane. In the protocol in ref. 21, this comprises steps 29–41, in particular step 40. Second, it is critical to the success of the experiment to wash the cellulose membranes carefully after the cleavage of the side chain protection groups. In ref. 21, this comprises steps 51–53. At this stage, remove all residual chemicals, wash with dichloromethane (five times for 3 min each time), dimethylformamide (five times for 3 min each time), ethanol (five times for 3 min each time) and an appropriate buffer such as 100 mM Tris–HCl, pH 7.3 (five times for 10 min each time). Note that any residual chemicals can interfere with the killing assay and non-peptide-related killing may occur. If peptides are obtained commercially or from another laboratory, inform them about these two points.
This protocol describes the screening of 100 peptides (excluding negative and positive control peptides) for antimicrobial activity against P. aeruginosa strain H1001 (ref. 9). Bac2A (RLARIVVIRVAR) was used as a positive control, and an unrelated peptide (GATPEDLNQKLS) was used as a negative control. As discussed above, we recommend first performing this protocol using only the positive and negative controls as well as randomly selected peptides from the screening set.
Materials
REAGENTS
-
2.1% (wt/vol) Difco Müller–Hinton (MH) broth sterilized by autoclaving (Becton-Dickson, Sparks, MD, cat. no. 275730)
-
2.1% (wt/vol) Difco MH broth and 1.5% agar (Becton-Dickson, Sparks, MD, cat. no. 281230), sterilized by autoclaving; transferred into Petri dishes (Fisher Scientific, Ottawa, Canada, cat. no. 0875712)
-
Tris (MP Biomedicals, Eschwege, Germany, cat. no. 819638)
-
Glucose (Sigma-Aldrich, Steinheim, Germany, cat. no. G7528-250G)
-
Endotoxin-free water (optional) (Sigma-Aldrich, Steinheim, Germany, cat. no. 2107-30ML)
-
Solution A (see REAGENT SETUP)
-
Solution B (see REAGENT SETUP)
Equipment
-
96-well polypropylene microtiter plates (Corning Inc., Corning, NY, cat. no. 3790)
Critical
Avoid tissue-culture-treated or polystyrene plates as these are strongly negatively charged and will non-specifically bind peptides.
-
96-well plates suitable for luminescence as indicated by the manufacturer, e.g., round-bottomed white and black 96-well plates from Perkin Elmer Life Science (Boston, MA, cat. no. 6005299)
-
Single hole punch (Staples, Canada)
-
Shaker, suitable for 96-well plates
-
Parafilm (Pechiney Plastic Packaging, Neenah, WI)
-
Sterile glass or plastic tubes with lid
-
Luminometer suitable for measuring luminescence in 96-well microtiter format, e.g., Tecan Spectra Fluor plus (Tecan US, Durham, NC)
-
12-channel multipipettor (e.g., VWR International, Edmonton, Canada, cat. no. 83009-710)
REAGENT SETUP
-
Solution A 100 mM Tris–HCl buffer, pH 7.3, 20 mM glucose.
-
Solution B 48 ml solution A and 1,920 μl of the microbial culture P. aeruginosa H1001 (fliC∷luxCDABE) at an OD600 of 0.35. The strain H1001 is available from us as part of our strain library, found at http://pseudomutant.pseudomonas.com.
Equipment setup
-
Advised settings for Tecan Spectra Fluor plus General measurement method: luminescence Plate definition file: GRE96ft.pdf
-
Measurement parameters Luminescence integration time: 1,500 ms Gain: 150 No kinetics
-
Shaking before measurement Duration: 10 s Mode: orbital Intensity: normal Settle time: 5 s
Procedure
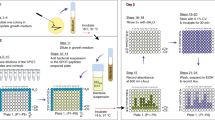
Preparation of microbial cultures
Timing Overnight
-
1
Pick a single colony of P. aeruginosa strain H1001 (ref. 9) from a freshly made MH agar plate and inoculate this colony into a sterile tube containing 3 ml MH. Alternatively, any other nutrient-rich medium can be used. To grow strains that contain a plasmid with an antibiotic resistance marker, use agar plates containing the corresponding antibiotic to ensure maintenance of the plasmid, but do not add antibiotic to the overnight culture or subsequent subcultures.
Pause point
Incubate the tube overnight in a shaker at 225 r.p.m. at 37 °C. This culture will be utilized in Step 10.
Preparation of the plates and isolation of peptide spots
Timing 60 min
-
2
Label all required 96-well plates, two polypropylene plates for storage of the peptides (label 'store 1' and 'store 2') and ten plates suitable for the luminescence assay (see Experimental design and Table 1). We recommend first testing the luminescence of the bacteria without any peptides to avoid wasting peptides in case the bacteria do not display any luminescence. Therefore, prepare one extra plate for testing the luminescence; this plate can be reused for additional tests. Label it 'test 1'.
-
3
Label all peptide spots, but not positive and negative control peptides, with consecutive numbers from 1 to 100 with a fine pencil. Label positive control 'P' and negative control 'N'.
Critical Step
Wear gloves to prevent contamination of the peptide spots with antimicrobial substances from the skin.
-
4
Punch out peptide spots and transfer each spot with tweezers into a separate well in the corresponding 96-well polypropylene plate ('store 1' and 'store 2'); peptides 1–10 will be transferred into row A, columns 2–11; peptides 11–20 will be transferred into row B, columns 2–11, and so on. Into each well of column 1 transfer a positive control peptide spot, and into each well of column 12 a negative control peptide spot, remembering to add negative and positive controls only when screening peptides are present in the same row.
Critical Step
Beware electrostatic interactions between the cellulose and 96-well plate that may cause cellulose spots to be ejected from the well. Before proceeding to Step 5, double-check that each peptide is in the corresponding well. The conclusions drawn from this assay depend on every peptide being correctly transferred into the corresponding well of the 96-well plates. Ideally, use a puncher that punches holes a little smaller than the diameter of the peptide spots. Carefully punch out the peptide spots so that their labeled centers lie right in the middle of the punched-out spot. If this is not done, the actual peptide concentration in the assay may decrease, leading to erroneous conclusions.
Releasing the peptides from the spots
Timing At least 2 h or overnight
-
5
Add 60 μl distilled and autoclaved water into each well. Alternatively, any non-toxic buffer can be used for solubilization of the peptides. Endotoxin-free water can be used for additional tissue culture experiments (e.g., cytotoxicity assays or assays to identify immunomodulatory effects). If peptide activity is not known, we recommend suspending the control peptides as well as the randomly selected peptides in different amounts of water/buffer, for example adding 50, 100 and 200 μl to separate spots of the same peptide (see Experimental design). The amount of water to be added depends on the activity level of the antimicrobial peptide and the types and density of amino functionalization of distinct cellulose membranes. Reported peptide densities vary between 0.05 and 1.9 μmol cm−2 (for a review see ref. 7). In our laboratory we use peptide spots with a density of 0.5–0.8 μmol cm−2, and each spot has an average area of approximately 0.25 cm2.
Critical Step
Peptide concentration is a critical parameter affecting the result of this assay. The optimal peptide concentration is one at which the researcher observes killing activity within the first dilutions (see Table 1) and no or weak killing activity at the highest dilution (see Table 1). In addition, you must account for the fact that the need to add enough liquid to the peptides to perform a second assay (an additional antimicrobial assay to confirm the data obtained or to determine cytotoxicity) might also influence the decision on the amount of liquid added.
-
6
Cover the two plates with a lid and seal the lids with Parafilm.
-
7
Set the plates on a microtiter plate shaker suitable for 96-well plates.
-
8
Shake for at least 2 h at room temperature (20–23 °C) (for fluorescence-labeled peptides cover plates with tinfoil).
Pause point
The plates can be shaken at room temperature overnight.
-
9
Use peptides directly for the assay or store at −20 °C.
Pause point
Peptides can be stored at −20 °C in the 96-well plate for several months (keep in the dark).
Determination of antimicrobial activity
-
10
To determine microbial activity using a bioluminescence assay (e.g., with strain H1001) choose option A. This is the most sensitive and fastest method. Alternatively, if no bioluminescent strain is available, use option B.
-
A
Microbes with a bioluminescence marker
-
i
Add 50 μl of the overnight culture into 4,950 μl MH broth in a tube.
-
ii
Incubate the tube in a shaker at 225 r.p.m. at 37 °C until the OD600 reaches 0.35 (approximately 2 h).
-
iii
Add 1,920 μl of the microbial culture into 48,000 μl solution A (see REAGENTS) to make solution B. This amount of solution B is required for ten complete 96-well plates. Use a sterile glass or plastic tube to prepare Solution B.
-
iv
Vortex solution B (see REAGENTS) gently for 2 min.
-
v
Transfer 50 μl of solution B into each of the 12 wells of row A of the 96-well plate labeled 'test 1'.
-
vi
Test the luminescence signal of the bacteria in these wells. If a luminescence signal is detected, follow the remainder of this protocol; otherwise stop at this point. At the equipment setting described, we normally measure more than 900 light units.
-
vii
Transfer 90 μl of solution B into each of the 12 wells of row A (numbered 1–12) of all ten 96-well plates suitable for bioluminescence measurement. Add 50 μl solution B into all other wells of all ten plates (into all other wells in rows B–H).
-
viii
Add 10 μl of peptide solution from plates 'store 1' and 'store 2' into the corresponding 12 wells of row A of all ten 96-well plates suitable for bioluminescence measurement (100 μl in total in this well). Peptides 1–10 and one positive and one negative control will be added to luminescence plate 1; peptides 11–20 and one positive and one negative control will be added to luminescence plate 2; and so on until all rows A of the ten plates are filled. To determine the true peptide concentration, add known concentrations of the positive control (e.g., solid-phase resin-synthesized Bac2A) to the two plates. Comparing analog luminescence data from cellulose-synthesized Bac2A with those from resin-synthesized Bac2A will allow the actual concentration of the cellulose-synthesized peptide to be estimated. If resin-synthesized Bac2A is not available, it is sufficient to determine relative activities.
-
ix
Store plates 'store 1' and 'store 2' at −20 °C.
-
x
Mix the contents of row A wells thoroughly and transfer (using a 12-channel multipipettor) 50 μl from each well of row A into each corresponding well of row B, e.g., 50 μl from A1 into B1. Mix solution in row B. The volume in each well of row B is now 100 μl and the peptide concentration one half of that in row A. Continue the stepwise dilution series, transferring 50 μl from one well to the next until row G, leaving row H untouched (this row will be used to obtain the 100% luminescence value for the untreated microbes). After mixing the solution in row G, discard 50 μl. Repeat this for all ten luminescence plates.
-
xi
Measure luminescence of the plates.
-
xii
Incubate the plate at 37 °C for 4 h.
-
xiii
Measure luminescence of all the plates. If most of the peptides cause luminescence to decline too slowly, select a later reading time; for example, measure luminescence after 8 h. Alternatively, if the peptide concentration proves too low, consult the troubleshooting section.
-
xiv
Add 70 μl MH medium into each well. Cover the ten plates with lids and seal the lids with Parafilm.
Pause point
Incubate overnight at 37 °C.
-
xv
After 16–20 h incubation time or when satisfactory growth is obtained (the wells show a visible turbidity), determine visually in which wells no microbial growth has occurred. Alternatively, transfer the contents of each well into a clear-bottomed 96-well plate and read OD600 of the entire plate (you can use a clear-bottomed luminescence plate from the start, but this type of plate is more expensive). By comparing the luminescence data with these growth data, it is possible to identify those peptides whose activity is strongly influenced by cation concentration (e.g., magnesium) as well as those peptides with killing abilities over a longer period of time (see ANTICIPATED RESULTS).
Timing 6 h
-
i
-
B
Alternative assay (microbes without a bioluminescence marker)
-
i
Dilute the overnight culture using MH to approximately 1 × 106 colony forming units per ml.
-
ii
Add 50 μl of peptide solution from plates 'store 1' and 'store 2' into the corresponding row A of all ten polypropylene 96-well plates. Peptides 1–10 and one positive and one negative control will be added to polypropylene plate 1; peptides 11–20 and one positive and one negative control will be added to polypropylene plate 2, and so on until all rows A of the ten plates are filled. To determine the true peptide concentration, add known concentrations of the positive control (e.g., solid-phase resin-synthesized Bac2A) to the two plates. If Bac2A is not available, it is sufficient to determine relative activities.
-
iii
To all other wells add 25 μl autoclaved dH2O. Perform a stepwise dilution series (transferring 25 μl from row A into row B and so on until row G, leaving row H untouched) using a 12-channel multipipettor. After mixing the solution in row G discard 25 μl.
-
iv
Add 75 μl of diluted overnight cultures [prepared at Step 10B(i)] into all wells of the plate.
Pause point
Incubate overnight at 37 °C
-
v
After 16–20 h incubation time or when satisfactory growth has been obtained, determine visually in which wells no microbial growth has occurred. Alternatively, transfer the contents of each well into a clear-bottomed 96-well plate (you can use a clear-bottomed plate from the start, but this type of plate is more expensive) and read the OD600 of the entire plate.
Timing 2 h–overnight
-
i
-
A
Analysis of results
Timing 1 h
-
11
To analyze the bioluminescence results from a screen of large numbers of peptides follow option A; to analyze smaller numbers of peptides or to assess peptides with similar activities (e.g., substitution analysis experiments) follow option B. To analyze data from non-bioluminescent microbes, go to option C.
-
A
Option A
-
i
Use the last row of each plate (labeled row H) to determine the average value for the luminescence of the microbes only. Using this value, a numeric cut-off value can be chosen, e.g., a 50 or 90% decrease in luminescence.
-
ii
Add a column labeled concentration to the results table, and fill out the column. To determine the concentration, compare the activity of resin-synthesized Bac2A, at known concentrations [Step 10A(viii)], with that of Bac2A synthesized on cellulose. This allows one to judge the true peptide concentration (on the basis of the dilution of cellulose-synthesized peptide that gives the same decrease in luminescence as resin-synthesized Bac2A). If no resin-synthesized peptide is available, the highest concentration of each peptide is assigned a value of 1.0 and the doubling dilutions are assigned the values 0.5, 0.25, 0.125 and so on (see Table 2). In row H fill in 0. An example is given in Table 2.
Table 2 Example of a bioluminescence readout in a 96-well plate using Pseudomonas aeruginosa strain H1001. -
iii
In each row, start from the bottom and move upward to assess the concentration that represents the first numeric value that is below the chosen cut-off value. This concentration represents the inhibitory concentration (IC). Table 2 shows an example of luminescence values and the corresponding concentrations where the cut-off has been reached.
Critical Step
If different peptides give quite similar cut-off values use the more sensitive analysis in option B.
-
iv
If the positive control peptide shows the same killing activity on all plates, present IC values as concentrations. If the positive control shows different killing activity between plates, use relative values referring to the positive control.
-
v
Add a column labeled concentration to the results table obtained from Step 10A(xv), and fill out the column, starting with 1 and lowering the concentration to 0.5 (according to the stepwise dilution) for the next step. In row H fill in 0. The ICOV (obtained after overnight/16–20 h incubation) is defined as the lowest concentration that inhibits visible growth as observed with the naked eye. Comparing the IC value with the ICOV value will help identify peptides that are strongly influenced by cation concentrations as well as those displaying killing abilities over a longer period of time (see ANTICIPATED RESULTS). Compare first the IC and ICOV values of the positive controls in the same plate, and then compare peptides only in relation to the positive control. Peptides that are influenced strongly by cations or suppress luminescence only over a short period of time will have a lower IC/ICOV ratio than the positive control.
-
i
-
B
Option B
-
i
To characterize similar peptides, use a more sensitive analysis method (IC50 values). IC50 values take all values of the dilution series into account, reflecting the response of each peptide in more detail. A relative or the true peptide concentration needs to be determined. To determine that concentration, use, e.g. known concentrations of the positive control Bac2A that has been solid-phase resin-synthesized [see Step 11A(viii)]. Comparing the activity of this positive control with that of the corresponding control peptide synthesized on cellulose allows one to judge the true peptide concentration (on the basis of the dilution of cellulose-synthesized peptide that gives the same decrease in luminescence as resin-synthesized Bac2A). If no resin-synthesized peptide is available, the highest concentration of each peptide is assigned a value of 1.0 and the doubling dilutions are assigned the values 0.5, 0.25, 0.125 and so on (see Table 2).
-
ii
Calculate the maximum luminescence value across the entire data set (Lmax). Each individual luminescence value (L) from each row is subtracted from this Lmax value. The log values of the peptide concentration are determined and plotted against the decrease in luminescence for each corresponding data point (Lmax−L). The IC50 value, where half the maximal rate of killing is observed, is determined as half of Lmax minus the lowest subtracted luminescence value for the positive control peptide on the respective plate.
-
iii
Perform linear regression for the graphs obtained for each peptide and, as long as the r2 (correlation coefficient) value is greater than 0.9, the concentration at the half maximal killing value is determined (see Table 3). Occasionally, the plot of subtracted luminescence versus log concentration may demonstrate saturation of the curve at high or low concentrations. In these instances, these values in the saturation area should be eliminated from the calculation of the linear regression and the interpolation procedure can be repeated as long as the resulting r2 value is greater than 0.9.
Table 3 Relative antimicrobial activities from transformed bioluminescence values.
-
i
-
C
Option C
-
i
Determine the true peptide concentration. Comparing the activity of a resin-synthesized Bac2A at known concentrations [Step 10B(ii)] with that of the corresponding control peptide synthesized on cellulose allows one to judge the true peptide concentration (on the basis of the dilution of cellulose-synthesized peptide that gives the same inhibition of growth as resin-synthesized Bac2A). If no resin-synthesized peptide is available, the highest concentration of each peptide is assigned a value of 1.0 and the doubling dilutions are assigned the values 0.5, 0.25, 0.125 and so on (see Table 2). In row H fill in 0.
-
ii
In each single row start from the top and move downward to determine the lowest concentration where no bacterial growth is observed. This represents the IC for the non-luminescence assay (ICNL).
-
iii
When the positive control peptide shows the same killing activity on all plates, ICNL values can be presented as concentrations. Conversely, if the positive control is different between plates, we recommend using relative values referring to the positive control.
-
i
-
A
Troubleshooting
Troubleshooting advice can be found in Table 4.
Timing
First day (Steps 1–8): approximately 2 h
Second day: approximately 6 h
Third day: approximately 1–2 h
ANTICIPATED RESULTS
Once an appropriate peptide concentration has been found (using the control spots), the antimicrobial assay is easy to perform. For one person it is possible to screen 100–300 novel peptides per day, working on a 3-d cycle. In combination with peptide synthesis on cellulose arrays, this makes it feasible to adopt systematic approaches, such as a substitution analysis or length analysis, to investigate the primary sequence determinants of peptide antimicrobial activity. An illustrative substitution analysis of the antimicrobial peptide Bac2A, and the activity of the derivative peptides against P. aeruginosa strain H1001, is shown in Figure 1. Peptide libraries can be used to screen thousands of individual peptides against a microbe of interest. Peptide array synthesis on cellulose also provides the possibility of designing combinatorial peptide libraries and screening millions of peptide mixtures.
The first two columns give the position (indicated as the row number) and the one-letter code sequence of the original peptide Bac2A-NH2. The second and third rows give, respectively, the column number and the amino acids substituted at each amino acid position. Thus, for example, the peptide in the upper left-hand corner (column 1, row 1) is ALRARIVVIRVAR, and the next peptide in row 1 is CLRARIVVIRVAR. The next peptide in row 2, column 1, is RARARIVVIRVAR; then the parent peptide Bac2A-NH2 appears in row 3, column 1 (RLARIVVIRVAR); the peptide in row 4, column 2 is RRACIVVIRVAR and so on. The results within each box represent the proxy IC50 value, determined by treatment of the lux reporter strain H1001 with peptide for 4 h. Results are color-coded: black represents superior activity to the parent peptide Bac2A; dark gray with white lettering represents equivalent activity to the parent peptide; light gray with black lettering represents inferior activity to the parent peptide Bac2A; white represents very little activity.
Microbes that express an energy-dependent bacterial luciferase constitute a sensitive tool for studying killing by antimicrobial peptides in a time-dependent manner (Fig. 2). The effect of peptides on cellular energy levels can occur in a time range from seconds to 30 min. In addition to assessing concentration-dependent killing or bacteriostatic effects after 20 h (similar to classical MIC studies), a real-time measurement detecting the decrease in cellular energization of the microbes can be performed. 'Classical' kill curves and the curves based on the inhibition of luminescence overlap well for most peptides (e.g., Fig. 3). Most peptides behave in a consistent manner; however, in screening thousands of different peptides, we discovered that relying on luminescence data gathered in Tris buffer, where the concentration of cations is low, might lead to wrong conclusions for peptides whose antimicrobial activity is strongly influenced by cations. The MIC value of such peptides observed in MH broth is much higher than expected. Therefore adding MH broth after the luminescence measurement is complete will indicate for which peptides the antimicrobial activity is strongly influenced by cations. In addition, a few peptides also showed a killing effect on the bacteria (decrease in luminescence), but after 8–10 h the bacteria recovered and the luminescence returned to the level for untreated bacteria. To filter out such peptides, we add MH broth after the luminescence measurement is completed and incubate for 16–20 h or until satisfactory growth is obtained.
Light-producing wells appear white. Note that at this plate in column 12 a peptide for screening was added, not a negative control (compare Table 1).
Bac2A was used at threefold the minimal inhibitory concentration for both methods; the triangles indicate the decrease in luminescence, and the squares indicate surviving bacterial colony counts. The circles indicate the lux assay without peptide. Both assays were performed in 100 mM Tris buffer pH 7.3 (containing 20 mM glucose in the case of the lux assay) with a starting concentration of between 2 × 106 and 2 × 107 colony forming units per ml.
We have described here a procedure for determining relative activity of peptides suitable for analysis using bioluminescence to screen large numbers of peptides. Tables 2 and 3 show an example of luminescence values and the corresponding concentrations where the cut-off is reached. In situations where the positive control is different between plates, we recommend using relative values referring to the positive control.
Microbes without bioluminescence can also be used. The results of the screen are not as sensitive as those obtained using luminescence, and thus will have inferior discriminatory ability. However, any strain of interest can be used and a luminescence cassette is not required. The readout of the plate is similar to the classical MIC determination; a schematic example is given in Figure 4.
Note that this plate is set up as described in Table 1: in column 1 a positive control was added and in column 12 a negative control was added. The wells without shading represent wells with no microbial growth, and gray shading indicates residual bacterial growth. In this example the peptide in column 6 has no antimicrobial activity against the tested microbe, whereas the peptide in column 2 is most effective (provided that the starting concentrations of all substances were equal). This results need to be verified using peptides of known concentration and purity (synthesized on resin).
In general, all results obtained from peptides synthesized on cellulose must be considered only preliminary. Peptide sequences of interest have to be re-synthesized using the classical synthesis on resin, with the isolated peptides being first purified and tested for biological activity. In our experience, similar peptides can show dramatic differences in activity or no changes at all. These changes are not predictable as they almost certainly reflect the 3D structure of the peptide relative to its target, and predicting the 3D structure of peptides, especially ones that enter membranes and change their structure, is as yet not possible. Thus, scrambling a peptide sequence can result in no activity or similar or higher activity compared to the parent peptide10. However, the screening method described here, when combined with the SPOT peptide array synthesis, is a very powerful tool for obtaining optimized peptides. For example, screening of a semi-random peptide library consisting of 1,000 peptides led to antimicrobial peptides with a success-rate of 30%, where 3% showed strong activity and 0.3% superior broad-spectrum activity (K. Hilpert, R. Volkmer-Engert and R.E.W. Hancock, unpublished results).
References
CDC. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004 Am. J. Infect. Control. 32, 470–485 (2004).
Jenssen, H., Hamill, P. & Hancock, R.E.W. Peptide antimicrobial agents. Clin. Microbiol. Rev. 19, 491–511 (2006).
Pini, A. et al. Antimicrobial activity of novel dendrimeric peptides obtained by phage display selection and rational modification. Antimicrob. Agents Chemother. 49, 2665–2672 (2005).
Jose, J., Betscheider, D. & Zangen, D. Bacterial surface display library screening by target enzyme labeling: identification of new human cathepsin G inhibitors. Anal. Biochem. 346, 258–267 (2005).
Xie, Q. et al. In vitro system for high-throughput screening of random peptide libraries for antimicrobial peptides that recognize bacterial membranes. J. Pept. Sci. 12, 643–652 (2006).
Frank, R. Spot synthesis: an easy technique for positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 48, 9217–9232 (1992).
Hilpert, K., Winkler, D.F.H. & Hancock, R.E.W. Cellulose-bound peptide arrays: preparation and applications. Biotech. Genetic Engineering Rev 24, 31–106 (2007).
Hilpert, K., Volkmer-Engert, R., Walter, T. & Hancock, R.E.W. High-throughput generation of small antibacterial peptides with improved activity. Nat. Biotechnol. 23, 1008–1012 (2005).
Lewenza, S. et al. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res. 15, 583–589 (2005).
Hilpert, K. et al. Sequence requirements and an optimization strategy for short antimicrobial peptides. Chem. Biol. 13, 1101–1107 (2006).
Blondell, S.E., Takahashi, E., Weber, P.A. & Houghton, R.A. Identification of antimicrobial peptides by using combinatorial libraries made up of unnatural amino acids. Antimicrob. Agents Chemother. 38, 2280–2286 (1994).
Naveh, A., Potasman, I., Bassan, H. & Ulitzur, S. A new rapid and sensitive bioluminescence assay for antibiotics that inhibit protein synthesis. J. Appl. Bacteriol. 56, 457–463 (1984).
Wheat, P.F., Spencer, R.C. & Hastings, J.G. A novel luminometer for rapid antimicrobial susceptibility tests on Gram-positive cocci by ATP bioluminescence. J. Med. Microbiol. 29, 277–282 (1989).
Shawar, R.M. et al. Rapid screening of natural products for antimycobacterial activity by using luciferase-expressing strains of Mycobacterium bovis BCG and Mycobacterium intracellulare . Antimicrob. Agents Chemother. 41, 570–574 (1997).
Loeliger, B. et al. Antibiotic-dependent correlation between drug-induced killing and loss of luminescence in Streptococcus gordonii expressing luciferase. Microb. Drug Resist. 9, 123–131 (2003).
Travis, S.M. et al. Bactericidal activity of mammalian cathelicidin-derived peptides. Infect. Immun. 68, 2748–2755 (2000).
Winson, M.K. et al. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol. Lett. 163, 193–202 (1998).
Kramer, A. et al. Spot-synthesis: observations and optimizations. J. Pept. Res. 54, 319–327 (1999).
Landgraf, C. et al. Protein interaction networks by proteome peptide scanning. PLoS Biol. 2, E14 (2004).
Weiser, A.A. et al. SPOT synthesis: reliability of array-based measurement of peptide binding affinity. Anal. Biochem. 342, 300–311 (2005).
Hilpert, K., Winkler, D.F.H. & Hancock, R.E.W. Peptide arrays on cellulose support: SPOT synthesis—a time and cost efficient method for synthesis of large numbers of peptides in a parallel and addressable fashion. Nat. Protoc. 2, 1333–1349 (2007).
Acknowledgements
We acknowledge the financial assistance of the Applied Food and Materials Network and the Canadian Institutes of Health Research. R.E.W.H. was supported by a Canada Research Chair award. K.H. was supported by a fellowship from the Canadian Institutes of Health Research. We thank R.A. Klady for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The method described in this paper is included in a US patent submission by the University of British Columbia.
Rights and permissions
About this article
Cite this article
Hilpert, K., Hancock, R. Use of luminescent bacteria for rapid screening and characterization of short cationic antimicrobial peptides synthesized on cellulose using peptide array technology. Nat Protoc 2, 1652–1660 (2007). https://doi.org/10.1038/nprot.2007.203
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.203
This article is cited by
-
In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design
Nature Communications (2018)
-
A short artificial antimicrobial peptide shows potential to prevent or treat bone infections
Scientific Reports (2017)
-
The rumen microbiome: an underexplored resource for novel antimicrobial discovery
npj Biofilms and Microbiomes (2017)
-
Optimization of oncocin for antibacterial activity using a SPOT synthesis approach: extending the pathogen spectrum to Staphylococcus aureus
Amino Acids (2016)
-
Evaluation of anti-PAIIL lectin hen yolk antibody as an agent inhibiting Pseudomonas aeruginosa adherence to epithelial cells
Monatshefte für Chemie - Chemical Monthly (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.