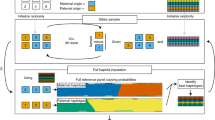
Abstract
The tagged microarray marker (TAM) method allows high-throughput differentiation between predicted alternative PCR products. Typically, the method is used as a molecular marker approach to determining the allelic states of single nucleotide polymorphisms (SNPs) or insertion–deletion (indel) alleles at genomic loci in multiple individuals. Biotin-labeled PCR products are spotted, unpurified, onto a streptavidin-coated glass slide and the alternative products are differentiated by hybridization to fluorescent detector oligonucleotides that recognize corresponding allele-specific tags on the PCR primers. The main attractions of this method are its high throughput (thousands of PCRs are analyzed per slide), flexibility of scoring (any combination, from a single marker in thousands of samples to thousands of markers in a single sample, can be analyzed) and flexibility of scale (any experimental scale, from a small lab setting up to a large project). This protocol describes an experiment involving 3,072 PCRs scored on a slide. The whole process from the start of PCR setup to receiving the data spreadsheet takes 2 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Wang, D.G. et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280, 1077–1082 (1998).
Schena, M., Shalon, D., Davis, R.W. & Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470 (1995).
Jenkins, S. & Gibson, N. High throughput SNP genotyping. Comp. Funct. Genomics 3, 57–66 (2002).
Hirschhorn, J.N. et al. SBE-TAGS: An array-based method for efficient single-nucleotide polymorphism. Proc. Natl. Acad. Sci. USA 97, 12164–12169 (2000).
Jaccoud, D., Peng, K., Feinstein, D. & Kilian, A. Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res. 29, e25 (2001).
Pastinen, T. et al. A system for specific, high-throughput genotyping by allele-specific primer extension on microarrays. Genome Res. 10, 1031–1042 (2000).
Guo, Z., Gatterman, M.S., Hood, L., Hansen, J.A. & Petersdorf, E.W. Oligonucleotide arrays for high-throughput SNPs detection in the MHC class I genes: HLA-B as a model system. Genome Res. 12, 447–457 (2002).
Lindroos, K., Sigurdsson, S., Johansson, K., Ronnblom, L. & Syvanen, A.-C. Multiplex SNP genotyping in pooled DNA samples by a four-colour microarray system. Nucleic Acids Res. 30, e70 (2002).
Engle, L.J., Simpson, C.L. & Landers, J.E. Using high-throughput SNP technologies to study cancer. Oncogene 25, 1594–1601 (2006).
Elder, J.K. et al. Antisense oligonucleotide scanning arrays. in DNA Microarrays: A Practical Approach (ed. Schena, M.) 77–98 (IRL Press, Oxford, UK, 1999).
Fan, J.B. et al. Highly parallel SNP genotyping. Cold Spring Harb. Symp. Quant. Biol. 68, 69–78 (2003).
Flavell, A.J. et al. A microarray-based high throughput molecular marker genotyping method: the tagged microrray (TAM) marker approach. Nucleic Acids Res. 31, e115 (2003).
Rust, S., Funke, H. & Assmann, G. Mutagenically separated PCR (MS-PCR): a highly specific one step procedure for easy mutation detection. Nucleic Acids Res. 21, 3623–3629 (1993).
Newton, C.R. et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 17, 2503–2516 (1989).
Kwok, S. et al. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 18, 999–1005 (1990).
Ye, S., Dhillon, S., Ke, X., Collins, A.R. & Day, I.N.M. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 29, e88 (2001).
Hayashi, K., Hashimoto, N., Daigen, M. & Ashikawa, I. Development of PCR-based SNP markers for rice blast resistance genes at the Piz locus. Theor. Appl. Genet. 108, 1212–1220 (2004).
Flavell, A.J., Knox, M.R., Pearce, S.R. & Ellis, T.H.N. Retrotransposon-based insertion polymorphisms (RBIP) for high throughput marker analysis. Plant J. 16, 643–650 (1998).
Nelson, S. & Denny, C.F. Representational difference analysis and microarray hybridization for efficient cloning and screening of differentially expressed genes. in DNA Microarrays: A Practical Approach (ed. Schena, M.) 77–98 (IRL Press, Oxford, UK, 1999).
Acknowledgements
This method could not have been developed without intellectual input from Pete Isaac and Noel Ellis. We are grateful to Mike Ferguson, without whom we would not have had easy access to microarray facilities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Jing, R., Bolshakov, V. & Flavell, A. The tagged microarray marker (TAM) method for high-throughput detection of single nucleotide and indel polymorphisms. Nat Protoc 2, 168–177 (2007). https://doi.org/10.1038/nprot.2006.408
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.408
This article is cited by
-
Genetic diversity in European Pisum germplasm collections
Theoretical and Applied Genetics (2012)
-
Analysis of plant diversity with retrotransposon-based molecular markers
Heredity (2011)
-
The genetic diversity and evolution of field pea (Pisum) studied by high throughput retrotransposon based insertion polymorphism (RBIP) marker analysis
BMC Evolutionary Biology (2010)
-
Temporal dynamics of receptor-induced apoptosis in an affinity microdevice
Analytical and Bioanalytical Chemistry (2010)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.