Abstract
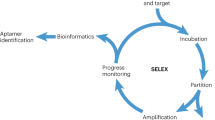
Aptamers are typically selected from libraries of random DNA (or RNA) sequences through systematic evolution of ligands by exponential enrichment (SELEX), which involves several rounds of alternating steps of partitioning of candidate oligonucleotides and their PCR amplification. Here we describe a protocol for non-SELEX selection of aptamers — a process that involves repetitive steps of partitioning with no amplification between them. Non-equilibrium capillary electrophoresis of equilibrium mixtures (NECEEM), which is a highly efficient affinity method, is used for partitioning. NECEEM also facilitates monitoring of bulk affinity of enriched libraries at every step of partitioning and screening of individual clones for their affinity to the target. NECEEM allows all clones to be screened prior to sequencing, so that only clones with suitable binding parameters are sequenced. The entire protocol can be completed in 1 wk, whereas conventional SELEX protocols take several weeks even in a specialized industrial facility.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Wilson, D.S. & Szostak, J.W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 68, 611–647 (1999).
Famulok, M., Mayer, G. & Blind, M. Nucleic acid aptamers — from selection in vitro to applications in vivo . Acc. Chem. Res. 33, 591–599 (2000).
Rimmele, M. Nucleic acid aptamers as tools and drugs: recent developments. Chembiochem 4, 963–971 (2003).
Jayasena, S.D. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 45, 1628–1650 (1999).
Lee, J.F., Stovall, G.M. & Ellington, A.D. Aptamer therapeutics advance. Curr. Opin. Chem. Biol. 10, 282–289 (2006).
Bock, C. et al. Photoaptamer arrays applied to multiplexed proteomic analysis. Proteomics 4, 609–618 (2004).
Surugiu-Warnmark, I., Warnmark, A., Toresson, G., Gustafsson, J.A. & Bulow, L. Selection of DNA aptamers against rat liver X receptors. Biochem. Biophys. Res. Commun. 332, 512–517 (2005).
Pestourie, C., Tavitian, B. & Duconge, F. Aptamers against extracellular targets for in vivo applications. Biochimie 87, 921–930 (2005).
Davidson, E.A. & Ellington, A.D. Engineering regulatory RNAs. Trends Biotechnol. 23, 109–112 (2005).
Mayer, G. & Jenne, A. Aptamers in research and drug development. BioDrugs 18, 351–359 (2004).
Nutiu, R., Yu, J.M. & Li, Y. Signaling aptamers for monitoring enzymatic activity and for inhibitor screening. Chembiochem 5, 1139–1144 (2004).
Famulok, M. & Mayer, G. Intramers and aptamers: applications in protein–function analyses and potential for drug screening. Chembiochem 6, 19–26 (2005).
Ulrich, H., Martins, A.H. & Pesquero, J.B. RNA and DNA aptamers in cytomics analysis. Cytometry A 59, 220–231 (2004).
Stojanovic, M.N. & Kolpashchikov, D.M. Modular aptameric sensors. J. Am. Chem. Soc. 126, 9266–9270 (2004).
Nimjee, S.M., Rusconi, C.P. & Sullenger, B.A. Aptamers: an emerging class of therapeutics. Annu. Rev. Med. 56, 555–583 (2005).
Patil, S.D., Rhodes, D.G. & Burgess, D.J. DNA-based therapeutics and DNA delivery systems: a comprehensive review. AAPS J. 7, 61–77 (2005).
Lee, J.F., Hesselberth, J.R., Meyers, L.A. & Ellington, A.D. Aptamer database. Nucleic Acids Res. 32, D95–D100 (2004).
Ellington, A. & Szostak, J. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
O'Connell, D. et al. Calcium-dependent oligonucleotide antagonists against L-selectin. Proc. Natl. Acad. Sci. USA 93, 5883–5887 (1996).
Ulrich, H. et al. In vitro selection of RNA molecules that displace cocaine from the membrane-bound nicotinic acetylcholine receptor. Proc. Natl. Acad. Sci. USA 95, 14051–14056 (1998).
Burke, D.H., Scates, L., Andrews, K. & Gold, L. Bent pseudoknots and novel RNA inhibitors of type 1 human immunodeficiency virus (HIV-1) reverse transcriptase. J. Mol. Biol. 264, 650–666 (1996).
Drolet, D.W., Jenison, R.D., Smith, D.E., Pratt, D. & Hicke, B. A high throughput platform for systematic evolution of ligands by exponential enrichment (SELEX). Comb. Chem. High Throughput Screen. 2, 271–278 (1999).
Beutel, B.A. & Gold, L. In vitro evolution of intrinsically bent DNA. J. Mol. Biol. 228, 803–812 (1992).
Ciesiolka, J. et al. Affinity selection-amplification from randomized ribooligonucleotide pools. Methods Enzymol. 267, 315–335 (1996).
Irvine, D., Tuerk, C. & Gold, L. SELEXION. Systematic evolution of ligands by exponential enrichment with integrated optimization by non-linear analysis. J. Mol. Biol. 222, 739–761 (1991).
Vant-Hull, B., Antonio, P.-B., Davis, R.H. & Gold, L. The mathematics of SELEX against complex targets. J. Mol. Biol. 278, 579–597 (1998).
Schneider, D., Gold, L. & Platt, T. Selective enrichment of RNA species for tight binding to Escherichia coli rho factor. FASEB J. 7, 201–207 (1993).
Chen, H. & Gold, L. Selection of high-affinity RNA ligands to reverse transcriptase: inhibition of cDNA synthesis and RNase H activity. Biochemistry 33, 8746–8756 (1994).
Tian, Y. et al. Dissecting protein:protein interactions between transcription factors with an RNA aptamer. RNA 1, 317–326 (1995).
Petrov, A., Okhonin, V., Berezovski, M. & Krylov, S.N. Kinetic capillary electrophoresis (KCE): a conceptual platform for kinetic homogeneous affinity methods. J. Am. Chem. Soc. 127, 17104–17110 (2005).
Heegaard, N.H.H. & Robey, F.A. Use of capillary zone electrophoresis to evaluate the binding of anionic carbohydrates to synthetic peptides derived from human serum amyloid P component. Anal. Chem. 64, 2479–2482 (1992).
Chu, Y.H., Avila, L.Z., Biebuyck, H.A. & Whitesides, G.M. Use of affinity capillary electrophoresis to measure binding constants of ligands to proteins. J. Med. Chem. 35, 2915–2917 (1992).
Mendonsa, S.D. & Bowser, M.T. In vitro evolution of functional DNA using capillary electrophoresis. J. Am. Chem. Soc. 126, 20–21 (2004).
Mosing, R.K., Mendonsa, S.D. & Bowser, M.T. Capillary electrophoresis-SELEX selection of aptamers with affinity for HIV-1 reverse transcriptase. Anal. Chem. 77, 6107–6112 (2005).
Berezovski, M. et al. Nonequilibrium capillary electrophoresis of equilibrium mixtures: a universal tool for development of aptamers. J. Am. Chem. Soc. 127, 3165–3171 (2005).
Drabovich, A.P., Berezovski, M., Okhonin, V. & Krylov, S.N. Selection of smart aptamers by methods of kinetic capillary electrophoresis. Anal. Chem. 78, 3171–3178 (2006).
Berezovski, M., Musheev, M., Drabovich, A. & Krylov, S.N. Non-SELEX selection of aptamers. J. Am. Chem. Soc. 128, 1410–1411 (2006).
Drabovich, A., Berezovski, M. & Krylov, S.N. Selection of smart aptamers by equilibrium capillary electrophoresis of equilibrium mixtures (ECEEM). J. Am. Chem. Soc. 127, 11224–11225 (2005).
Berezovski, M. & Krylov, S.N. Nonequilibrium capillary electrophoresis of equilibrium mixtures — a single experiment reveals equilibrium and kinetic parameters of protein–DNA interactions. J. Am. Chem. Soc. 124, 13764–13765 (2002).
Berezovski, M. & Krylov, S.N. Thermochemistry of protein–DNA interaction studied with temperature-controlled nonequilibrium capillary electrophoresis of equilibrium mixtures. Anal. Chem. 77, 1526–1529 (2005).
Thiel, K. Oligo oligarchy — the surprisingly small world of aptamers. Nat. Biotechnol. 22, 649–651 (2004).
Kanna, M.W., Rozenman, M.M., Sakurai, K., Snyder, T.M. & Liu, D.R. Reaction discovery enabled by DNA-templated synthesis and in vitro selection. Nature 431, 545–549 (2004).
Garner, Z.J. et al. DNA-templated synthesis and selection of a library of macrocycles. Science 305, 1601–1605 (2004).
Takahashi, T.T., Austin, R.J. & Roberts, R.W. mRNA display: ligand discovery, interaction analysis and beyond. Trends Biochem. Sci. 28, 159–165 (2003).
Don, R.H., Cox, P.T., Wainwright, B.J., Baker, K. & Mattick, J.S. 'Touchdown' PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19, 4008 (1991).
Musheev, M.U. & Krylov, S.N. Selection of aptamers by systematic evolution of ligands by exponential enrichment: addressing the polymerase chain reaction issue. Anal. Chim. Acta 564, 91–96 (2006).
Sanger, F., Nicklen, S. & Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 5463–5471 (1977).
Acknowledgements
This work was supported by the Natural Sciences and Engineering Research Council of Canada.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Berezovski, M., Musheev, M., Drabovich, A. et al. Non-SELEX: selection of aptamers without intermediate amplification of candidate oligonucleotides. Nat Protoc 1, 1359–1369 (2006). https://doi.org/10.1038/nprot.2006.200
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.200
This article is cited by
-
Selection and Characterization of Cell Surface Specific Aptamer and Development of Fluorescence Assay for Detection of Shigella flexneri from Water Samples
Journal of Fluorescence (2021)
-
Competitive non-SELEX for the selective and rapid enrichment of DNA aptamers and its use in electrochemical aptasensor
Scientific Reports (2019)
-
Prospects in the use of aptamers for characterizing the structure and stability of bioactive proteins and peptides in food
Analytical and Bioanalytical Chemistry (2018)
-
Short-chained oligo(ethylene oxide)-functionalized gold nanoparticles: realization of significant protein resistance
Analytical and Bioanalytical Chemistry (2018)
-
Use of Capillary Electrophoresis to Study the Binding Interaction of Aptamers with Wild-Type, K103N, and Double Mutant (K103N/Y181C) HIV-1 RT
Applied Biochemistry and Biotechnology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.