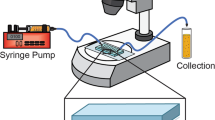
Abstract
The noninvasive character of NMR spectroscopy, combined with the sensitivity of the chemical shift, makes it ideally suited to investigate the conformation, binding events and dynamics of macromolecules inside living cells. These 'in-cell NMR' experiments involve labeling the macromolecule of interest with a nonradioactive but NMR-active isotope (15N or 13C). Cellular samples are prepared either by selectively overexpressing the protein in suitable cells (e.g., bacterial cells grown on isotopically labeled media), or by injecting isotopically labeled proteins directly into either cells or cell extracts. Here we provide detailed protocols for in-cell NMR experiments in the prokaryotic organism Escherichia coli, as well as eukaryotic cells and extracts employing Xenopus laevis oocytes or egg extracts. In-cell NMR samples with proteins overexpressed in E. coli can be produced within 13–14 h. Preparing Xenopus oocyte samples for in-cell NMR experiments takes 6–14 h depending on the oocyte preparation scheme and the injection method used.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Fiaux, J., Bertelsen, E.B., Horwich, A.L. & Wuthrich, K. NMR analysis of a 900K GroEL GroES complex. Nature 418, 207–211 (2002).
Williams, S.P., Haggie, P.M. & Brindle, K.M. 19F NMR measurements of the rotational mobility of proteins in vivo. Biophys. J. 72, 490–498 (1997).
Serber, Z. et al. Methyl groups as probes for proteins and complexes in in-cell NMR experiments. J. Am. Chem. Soc. 126, 7119–7125 (2004).
Brindle, K., Williams, S.P. & Boulton, M. 19F NMR detection of a fluorine-labelled enzyme in vivo. FEBS Lett. 255, 121–124 (1989).
Serber, Z., Ledwidge, R., Miller, S.M. & Dötsch, V. Evaluation of parameters critical to observing proteins inside living E. coli by in-cell NMR spectroscopy. J. Am. Chem. Soc. 123, 8895–8901 (2001).
Selenko, P., Serber, Z., Gadea, B., Ruderman, J. & Wagner, G. Quantitative NMR analysis of the protein G G1 domain in Xenopus laevis egg extracts and oocyte cells. Proc. Natl. Acad. Sci. USA 103, 11904–11909 (2006).
Dedmon, M.M., Patel, C.N., Young, G.B. & Pielak, G.J. FlgM gains structure in living cells. Proc. Natl. Acad. Sci. USA 99, 12681–12684 (2002).
McNulty, B.C. et al. Temperature-induced reversible conformational change in the first 100 residues of α-synuclein. Protein. Sci. 15, 602–609 (2006).
McNulty, B.C., Young, G.B. & Pielak, G.J. Macromolecular crowding in the Escherichia coli periplasm maintains α-synuclein disorder. J. Mol. Biol. 355, 893–897 (2006).
Burz, D.S., Dutta, K., Cowburn, D. & Shekhtman, A. Mapping structural interactions using in-cell NMR spectroscopy (STINT-NMR). Nat. Methods 3, 91–93 (2006).
Fesik, S.W. NMR structure-based drug design. J. Biomol. NMR 3, 261–269 (1993).
Shuker, S.B. et al. Discovering high-affinity ligands for proteins: SAR by NMR. Science 274, 1531–1534 (1996).
Hubbard, J.A., MacLachlan, L.K., King, G.W., Jones, J.J. & Fosberry, A.P. Nuclear magnetic resonance spectroscopy reveals the functional state of the signalling protein CheY in vivo in Escherichia coli. Mol. Microbiol. 49, 1191–1200 (2003).
Bryant, J.E., Lecomte, J.T.J., Lee, A.L., Young, G.B. & Pielak, G.J. Protein dynamics in living cells. Biochemistry 44, 9275–9279 (2005).
Reckel, S., Löhr, F. & Dötsch, V. In-cell NMR spectroscopy. Chembiochem 6, 1601–1606 (2005).
Xenopus laevis: practical uses in cell and molecular biology. Methods Cell Biol. 36, 1–718 (1991).
Scopes, R.K. Protein Purification: Principles and Practice 3rd edn. (Springer Press, New York, 2004).
Murray, A.W. Cell cycle extracts. Methods Cell Biol. 36, 581–605 (1991).
Schnizler, K., Kuster, M., Methfessel, C. & Fejtl, M. The Roboocyte: automated cDNA/mRNA injection and subsequent TEVC recording on Xenopus oocytes in 96-well microtiter plates. Receptors Channels 9, 41–48 (2003).
Acknowledgements
Z.S. acknowledges funding from the Beckman Foundation. P.S. acknowledges funding by a Human Science Frontier Project Organization long-term fellowship (LT00686/2004-C). S.R. acknowledges support from the Verband der Deutschen Chemischen Industrie. V.D. acknowledges support from the Center for Biomolecular Magnetic Resonance at the University of Frankfurt, Germany.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Serber, Z., Selenko, P., Hänsel, R. et al. Investigating macromolecules inside cultured and injected cells by in-cell NMR spectroscopy. Nat Protoc 1, 2701–2709 (2006). https://doi.org/10.1038/nprot.2006.181
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.181
This article is cited by
-
Monitoring and modelling the glutamine metabolic pathway: a review and future perspectives
Metabolomics (2023)
-
Investigation of higher-order RNA G-quadruplex structures in vitro and in living cells by 19F NMR spectroscopy
Nature Protocols (2018)
-
Caught in Action: Selecting Peptide Aptamers Against Intrinsically Disordered Proteins in Live Cells
Scientific Reports (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.