Abstract
Major depressive disorder is a chronic, remitting syndrome involving widely distributed circuits in the brain. Stable alterations in gene expression that contribute to structural and functional changes in multiple brain regions are implicated in the heterogeneity and pathogenesis of the illness. Epigenetic events that alter chromatin structure to regulate programs of gene expression have been associated with depression-related behavior, antidepressant action, and resistance to depression or ‘resilience’ in animal models, with increasing evidence for similar mechanisms occurring in postmortem brains of depressed humans. In this review, we discuss recent advances in our understanding of epigenetic contributions to depression, in particular the role of histone acetylation and methylation, which are revealing novel mechanistic insight into the syndrome that may aid in the development of novel targets for depression treatment.
Similar content being viewed by others
INTRODUCTION: WHY EPIGENETICS?
Major depressive disorder (MDD) is a chronic and debilitating syndrome that affects ∼16% of the US population (Kessler et al, 2003), and is ranked second globally in terms of disease burden (Murray and Lopez, 1997). It is evident from DSM-IV that major depression is a complex and heterogeneous disorder with a wide spectrum of symptoms including depressed mood, anhedonia, disturbed sleep, appetite, and energy, reduced concentration, excessive guilt, and suicidal thoughts (American Psychiatric Association, 2000). The etiology of the disorder is still not well understood. Meta-analysis of a large body of genetic epidemiological studies has revealed that heritability of major depression is between ∼31 and 42% (Sullivan et al, 2000); however, human GWAS studies have largely failed to produce reproducible gene loci that are contributing significantly to the disease (Bosker et al, 2011). Furthermore, the heritability of major depression is considerably lower compared with other psychiatric disorders such as schizophrenia and bipolar disorder, which show heritability rates of 70–80% (Kendler, 1983). The high discordance rate (50%) among monozygotic twins for MDD provides further evidence that genetics alone do not account fully for the illness (Fraga et al, 2005). Epidemiological evidence also links environmental factors, especially exposure to stressful life events, with an increased risk for depression (Hammen, 2005; Kessler, 1997). However, there is a remarkable individual variability in vulnerability to environmental stress, with many psychiatric disorders associated with a history of stress and most individuals showing normal outcomes despite stress exposure (Dudley et al, 2011). Owing to the lack of success of traditional genetic or environmental approaches alone to explain MDD, a recent prevalent theory in the field has focused on the interplay between genetic and environmental factors, specifically how susceptibility and resilience to disease is determined by the penetrance of highly complex genetic variation moderated by environmental cues and possibly stochastic or random events during brain development (Kendler, 1998; Lesch, 2011; Vialou et al, 2012).
The term ‘epigenetics’, coined by Waddington in the 1940s to address the central question of how a common genome generates numerous distinct cell types, originally referred to phenotypes resulting from interactions between genes and environment (Waddington, 1940, 1957; Waddington and International Union of Biological Sciences., 1968). Up to that point, the field had focused on the coupling between genetic and phenotypic variation. Waddington and others wanted to understand why frequently genetic and phenotypic variations appeared uncoupled (Jablonka and Lamb, 2002), one of the main issues facing the field of psychiatry today. The definition of epigenetics has changed over time, and the term now refers to the potentially heritable, but environmentally modifiable, regulation of genetic function and expression that is mediated through non-DNA-encoded mechanisms (Russo et al, 1996; Wu and Morris, 2001). Epigenetics has surfaced to the forefront as a mechanism by which environmental cues can be translated into precise and highly stable alterations in chromatin structure that ultimately lead to the persistent expression of altered gene programs.
Evidence derived mostly from a large body of research in animal models suggests that transcriptional dysregulation may underlie the behavioral manifestation of many psychiatric disorders including depression (Charney and Manji, 2004; Krishnan and Nestler, 2008). Accordingly, a recent hypothesis is that certain environmental factors hijack the brain's epigenetic machinery, best understood for its role in normal development and differentiation, and in combination with genetic predispositions produce many of the behavioral manifestations of these disorders (Peter and Akbarian, 2011; Renthal and Nestler, 2009). In fact, it has been demonstrated that, during the lifetime of monozygotic twins, there is a profound accumulation of epigenetic differences, likely because of different life experience as well as random events during development (Fraga et al, 2005). Numerous studies in animals have also shown that early life stress can leave persistent epigenetic marks in the genome, which alter gene expression and can influence neural and behavioral function through adulthood (McGowan et al, 2009, 2011; Murgatroyd et al, 2009; Weaver et al, 2004). In addition, recent evidence that monoamine oxidase inhibitors, a commonly used class of antidepressants, are potent inhibitors of the histone demethylase LSD1, and that valproate, an anticonvulsant used widely as a mood stabilizer, is also an inhibitor of histone deacetylases (HDACs), has given further impetus to the study of epigenetics mechanisms in the pathophysiology of depression (Harwood, 2003; Lee et al, 2006); although it is far from clear that these chromatin mechanisms account for the clinical actions of these drugs.
In this review, we first provide an overview of approaches ranging from animal models to human imaging studies, and then summarize results from those studies that have led to our understanding of the neural circuitry and associated molecular alterations in relevant brain regions implicated in the pathophysiology of depression. We then provide a very brief overview of epigenetic mechanisms, with a special focus on histone acetylation and methylation. Finally, we discuss the limited, albeit growing, number of studies in the field of epigenetics and depression, and relate how alterations in chromatin (both in terms of global/genome-wide changes as well as promoter-specific changes) may be linked to altered molecular patterns observed in depression.
ANIMAL MODELS OF DEPRESSION
The multitude of symptoms relevant to the diagnosis of human depression, and their varied prevalence and subjective nature, has posed significant challenges in the development of etiologically valid animal models of depression (Nestler and Hyman, 2010). One avenue upon which the field has capitalized is the observation that stressful life events, including trauma and emotional loss, are associated with heightened risk for depression in humans. The development of animal stress paradigms combined with the ability to objectively measure anhedonia and homeostatic symptoms in rodents have helped inform the neural circuitry and neuroadaptations underlying depression.
Animal models used to study depression can be broadly clustered into two categories; those employing acute stress procedures and those involving more chronic exposure to stressful stimuli. Acute stress paradigms such as the forced swim or tail suspension tests involve a short duration physical stress during which an animal's coping response is assessed by measuring time spent responding actively vs passively. Chronic stress paradigms involve prolonged exposure to either physical stressors (chronic mild or unpredictable stress; Willner, 2005) or bouts of social subordination (chronic social defeat stress; Berton et al, 2006). Such chronic stress assays, as well as chronic social isolation, produce anhedonia-like symptoms, characterized by a decrease in reward-related behaviors such as preferences for sucrose or high fat diets and social interaction (Berton et al, 2006; Wallace et al, 2009). Anhedonic phenotypes resulting from exposure to chronic stress paradigms are rarely seen following acute stress, suggesting the former is more successful at recapitulating behavioral features of human depression. Additionally, the anhedonia resulting from chronic stress can be effectively reversed by chronic but not acute treatment with current antidepressant medications (eg, Berton et al, 2006; Wallace et al, 2009), a treatment course comparable to that required in humans, whereas a single dose of antidepressant is sufficient to increase active responding in the forced swim and tail suspension tests (Cryan et al, 2002; Nestler et al, 2002). Together, these data suggest that chronic stress paradigms are more effective at modeling at least certain features or subtypes of the human depression syndrome, with acute studies providing insight into neuronal adaptations that regulate responses to stressful events.
CIRCUIT AND MOLECULAR MECHANISMS OF DEPRESSION
Postmortem and neuroimaging studies of depressed patients have revealed that human depression likely affects numerous regions of the brain, which may account for some of the complexity and heterogeneity of the disorder. Functional changes throughout the brain's cortical-striatal-limbic circuitry have been reported, suggesting altered cognition (prefrontal cortex (PFC) and hippocampus), emotional (amygdala) and reward (nucleus accumbens (NAc)) processing, and homeostatic and stress responses (eg, hypothalamic-pituitary-adrenal or HPA axis) during the course of depression. For example, studies have revealed decreased gray-matter volume in the PFC and hippocampus of depressed individuals (Drevets, 2001; Harrison, 2002; Sheline, 2003), hypermetabolism in the amygdala and frontal cortical regions (Drevets, 2003, 2007; Drevets et al, 2008; Fu et al, 2004), alterations in spine morphology (Christoffel et al, 2011) as well as decreased activation of the NAc in response to rewarding stimuli (Epstein et al, 2006), and hyperactivity of the HPA axis (Gold and Chrousos, 2002).
Molecular adaptations occurring throughout the cortical-striatal-limbic circuitry in human and animal models of depression are thought to underlie morphological and functional changes that contribute to the disease state (Figure 1). Human postmortem studies have revealed a decrease in brain-derived neurotrophic factor (BDNF) in the hippocampus of patients suffering from depression (Dwivedi et al, 2003). BDNF, the most active member of the neurotrophin family, regulates neuronal differentiation and growth (Benraiss et al, 2001; Pencea et al, 2001), and the decrease observed in depression may contribute to the reduction in hippocampal volume seen in depressed patients. Reduced levels of hippocampal BDNF are corroborated in both acute (Barrientos et al, 2003) and chronic (Nibuya et al, 1995) stress paradigms in animals. Evidence that antidepressant treatment in humans and animals causes an increase in hippocampal BDNF, and that antidepressant responses are lost upon BDNF knockout, suggests that this molecular adaptation is relevant for part of the symptomatology of depression (Coppell et al, 2003; Gervasoni et al, 2005; Monteggia et al, 2007; Nibuya et al, 1995; Shimizu et al, 2003; Xu et al, 2003). Interestingly, a similar hippocampal profile has been reported for several other growth factors, such as VGF, vascular endothelial growth factor (VEGF) and its receptor (VEGF type 2), in chronic stress models (Heine et al, 2005; Thakker et al, 2007; Warner-Schmidt and Duman, 2007). In addition to decreases in neuronal growth factors, both acute and chronic stress models in rodents produce an increase in hippocampal proinflammatory cytokine IL-1β and NF-κβ signaling, molecular adaptations that are linked to both depressive-like phenotypes and decreased hippocampal cell proliferation (Goshen et al, 2008; Johnson et al, 2005; Koo and Duman, 2008; Koo et al, 2010).
Circuitry with molecular adaptations overlaid onto different brain regions. Multiple brain regions are implicated in the pathophysiology of depression. Depicted here are examples of molecular adaptations occurring in the glutamatergic, dopaminergic (ventral tegmental area, VTA) and GABAergic systems in depressed humans and/or following chronic stress in rodent models. Changes highlighted in red are reversed by chronic treatment with antidepressants.
Depressed patients and animals subjected to chronic stress exhibit structural alterations in the PFC, including decreased PFC volume and dendritic retraction and spine loss (Cook and Wellman, 2004; Drevets, 2000, 2001; Goldwater et al, 2009; Liston et al, 2006). Depressed patients also display decreased levels of the NR2A and NR2B NMDA glutamate receptor subunits as well as a reduction in PSD-95, all of which are crucial for the structural integrity and function of dendritic spines (Gambrill and Barria, 2011). Chronic stress decreases levels of BDNF in the PFC (Fumagalli et al, 2004) and, similar to the hippocampus, levels are increased after treatment with antidepressants (Balu et al, 2009). Interestingly, neuropeptide Y, which antagonizes the actions of corticotrophin-releasing hormone (Giesbrecht et al, 2010; Heilig, 2004), is also decreased in the PFC of depressed patients and suicide victims (Caberlotto and Hurd, 2001; Widdowson et al, 1992). Adding complication to molecular approaches in treating depression is that molecular adaptations do not occur globally but are often region- and cell type-specific. For example, in contrast to what has been observed in the PFC, NR2A, and PSD-95 are both increased in the amygdala of depressed humans (Karolewicz et al, 2009).
The NAc, best established for its role in processing reward information, has been hypothesized to underlie symptoms of anhedonia in depressed patients (Nestler and Carlezon, 2006). Much of what is known about the molecular adaptations in the NAc in depression has been derived from chronic stress paradigms in rodents, particularly the chronic social defeat paradigm, with most key findings validated in the NAc of depressed humans. One advantage of chronic social defeat is that mice show phenotypically divergent responses, with some animals showing susceptibility and others demonstrating resilience (Krishnan et al, 2007), a phenomenon that could translate to differences in stress vulnerability seen in humans. BDNF levels are increased in the NAc after chronic social defeat in susceptible animals but not in their resilient counterparts (Berton et al, 2006; Krishnan et al, 2007). In addition, this selective increase in BDNF, demonstrated in the NAc of depressed humans as well, is associated with the activation of downstream signaling molecules such as phosphorylated akt thymoma viral oncogene and extracellular signal regulated kinase (Berton et al, 2006; Krishnan et al, 2007; Wilkinson et al, 2011). More recent evidence suggests a similar molecular distinction between susceptibility and resilience in the Wingless (WNT) signaling cascade in the NAc, with downregulation of WNT-dishevelled (DVL) signaling and associated upregulation of glycogen synthase kinase 3β signaling in susceptible mice but upregulation in resilient mice (Wilkinson et al, 2011). Downregulation of WNT-DVL signaling was corroborated in the NAc of human depressed patients examined postmortem. Changes in NAc spine morphology after chronic social defeat stress, specifically, an increase in stubby spines with smaller postsynaptic densities on NAc medium spiny neurons, are also specific to susceptible animals and absent in those demonstrating behavioral resilience (Christoffel et al, 2011). This adaptation appears to be mediated by induction of IkB kinase, which is a downstream target of BDNF and several other neurotrophic factors and a known regulator of spine morphology (Russo et al, 2009).
Observations in the chronic social defeat model, that mice with identical genetic backgrounds not only show differences in the development of anhedonic responses after exposure to the same environmental stress, but also show differences in the development of molecular adaptations in brain regions compromised in depression, suggests an interaction between genes and the environment that may be regulated through epigenetic mechanisms. Indeed, several regulators of chromatin structure are altered in human depression and animal models involving chronic stress, as will be seen below.
EPIGENETIC MECHANISMS
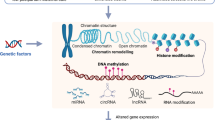
The eukaryotic genome contains a massive amount of genetic material (∼2 m of linear DNA) that has to be condensed into a microscopic nucleus (∼10 μm diameter). Chromatin, the histone, and non-histone proteins associating with DNA serves as an organizer of the genome by condensing the double-stranded DNA into multiple levels of higher order structures (Figure 2a). Chromatin also creates a barrier for transcription, replication, recombination, and repair, such that special molecular machinery must be recruited to reversibly disrupt and modify chromatin to balance between the effective storage vs functional utility of the genome. The structural unit of chromatin is the nucleosome, which is an octamer of core histones H2A, H2B, H3, and H4 around which 147 bp of DNA is superhelically wrapped. A fifth histone molecule, known as H1, serves as a linker that helps compact the nucleosomes into higher order structures (Allan et al, 1986).
Chromatin structure and histone modifications at N-terminal histone tails. (a) The eukaryotic genome is organized by wrapping DNA around histone octamers to form the basic units of chromatin, nucleosomes, which are then further organized and compacted into higher ordered structures. (b) The histone octamer consists of two copies each of H2A, H2B, H3, and H4. In addition to globular domains, they each have N-termini tails that protrude from the nucleosome while H2A also has a C-terminal tail. These tails can be posttranslationally modified, and all known mammalian acetylation and methylation modifications on lysine residues on each tail are highlighted. The molecules are drawn roughly to proportion to the size of the protein, although the number of residues shown is not meant to reflect the exact size of the N-terminal tails.
Each of the H2A, H2B, H3, and H4 histone molecules has a globular histone fold domain that accounts for ∼75% of its mass and contributes to the core nucleosome, whereas the remaining 25% is contributed by N-terminal tails that protrude from the core through the gyres and grooves of DNA molecules (Wolffe and Hayes, 1999). Numerous types of posttranslational modifications on these tails, such as acetylation and methylation, are reversibly added and removed by ‘writers’ and ‘erasers,’ respectively. Lysine residues on histone N-termini tails that are known to be acetylated or methylated are highlighted in Figure 2b. These modifications are important for chromatin structure and function, and can work by two non-exclusive mechanisms. First, chromatin structure and compaction can be altered because of steric or charge introduced by the modifications. Second, these posttranslational modifications serve as recognition sites that recruit machinery with ‘reader’ domains for these modifications such as the transcription complex, other histone modifiers, and ATP-dependent remodeling enzymes, to disrupt, rearrange, or reassemble chromatin. Strahl and Allis (2000) proposed the ‘histone code hypothesis,’ which postulates that the vast combinatorial possibilities of these distinct posttranslation modifications bring about distinct downstream events by recruiting distinct proteins that diferentially recognize combinations of these marks by their reader domains. This body of knowledge, in combination with studies demonstrating that environmental factors can recruit epigenetic machinery via several intracellular signaling pathways including in neuronal systems (Borrelli et al, 2008), suggest that the diversity of life experience can transduce differential epigenetic changes on top of existing genetic vulnerabilities to result in the complex and heterogeneous phenotypes we observe in psychiatric disorders including MDD.
An additional well-studied epigenetic mechanism is the methylation of the DNA molecule itself. Detailed mechanisms of DNA methylation as well as their relation to psychiatric disorders are reviewed elsewhere in this issue (see Fan 2012; Zhang et al, 2012, this issue). In addition, more detailed descriptions of the basic chromatin structure and its diverse regulation are also reviewed elsewhere (see Maze et al, 2012, this issue). Here, we focus on two of the best-studied posttranslational modifications of histones in depression models, histone lysine (K) acetylation and methylation.
Histone Acetylation
Allfrey et al were the first to demonstrate hyperacetylated histones in actively transcribed regions of chromatin and hypoacetylated histones in transcriptionally silent regions. The first direct link between transcriptional activation and histone acetylation was the discovery that coactivator complexes required for transcriptional activation function as histone acetyltransferases (HATs) (Brownell et al, 1996; Ogryzko et al, 1996), enzymes that add acetyl groups to histone tails. Later studies found that transcriptional co-repressor complexes contain HDACs (Alland et al, 1997; Taunton et al, 1996), enzymes that remove acetyl groups from histone tails. Histone acetylation states are highly dynamic, with estimates of half-lives in the range of minutes within transcriptionally active chromatin (Zhang and Nelson, 1988). Further evidence comes from studies demonstrating that lysine–to-glutamine mutations that mimic lysine acetylation in N-termini of H3 and H4 relieve the requirement for HAT activity in transcriptional activation (Zhang et al, 1998). Genome-wide analyses have found histone acetylation to be localized mainly within promoters but also at lower levels throughout the genome (Wang et al, 2008).
One hypothesis of the mechanism by which acetylation activates transcription is that it neutralizes the positive charge of the histone molecule, thereby disrupting interaction with the negatively charged DNA, leading to a more relaxed and accessible chromatin template (Choi and Howe, 2009; Kouzarides, 2007). Several studies suggest that although acetylated tails still make contact with DNA (Mutskov et al, 1998) they wrap DNA with reduced affinity, which results in more mobility with respect to the DNA surface compared with unmodified tails (Cary et al, 1982; Lutter et al, 1992). In addition to disruption of the nucleosome structure itself, histone acetylation has been found to disrupt the stability of higher order structure (Tse et al, 1998); however, the level of charge neutralization necessary to facilitate this destabilization is so low that other structural features must amplify the consequences of acetylation (Wolffe and Hayes, 1999). Two possibilities are steric structural alterations to the tail domain as well as recruitment of factors that bind acetylated histone residues (proteins with bromodomains, such as transcription factors) or exclusion of factors that do not bind acetylated residues.
HATs are broadly classified into type A and type B based on the functional localization in the nucleus and cytoplasm, respectively (Selvi and Kundu, 2009). Type A HATs are further divided into five classes based on the structural and functional differences (Grant and Berger, 1999). HDACs are mainly classified into four classes (Hildmann et al, 2007; Selvi and Kundu, 2009). Class I includes HDAC 1–3 and 8. These are located in the nucleus and are mainly involved in epigenetic regulation. Class II comprises HDAC4–7, 9, and 10. These HDACs contain domains that allow them to be shuttled between the nucleus and cytoplasm, and function to deacetylate both histones and cytoplasmic proteins. Class III HDACs, also known as the sirtuins, are distinguished by their NAD dependence, and are important regulators of metabolism and transcription through the deacetylation of numerous histone and non-histone substrates (Schwer and Verdin, 2008). Class IV refers to HDAC11 about which little is known (Gao et al, 2002). There is some residue specificity for HATs and HDACs, although targeting is quite broad compared with the histone methylation machinery; individual lysine residues appear to serve as substrates for many different HATs and HDACs and a single HAT or HDAC may act on multiple residues (Khan and Khan, 2010; Selvi and Kundu, 2009). There are also some overlapping functions of histone acetylation at the same residue; eg, transcriptional activation vs DNA repair. However, the targeting of HATs to a given residue is largely unknown, with the likely involvement of several associated proteins (Khan and Khan, 2010; Selvi and Kundu, 2009).
Histone Lysine Methylation
Although histone methylation can occur at lysine, arginine, or histidine residues (Izzo and Schneider, 2010), we focus here on lysine methylation, the only form implicated to date in depression. The function of lysine methylation depends on the residue as well as the valence state of the methylation (Berger, 2007). A lysine residue can be mono-, di-, or trimethylated. Site- and state-specific lysine methylation is catalyzed by a group of lysine methyltransferases (KMT). With the exception of DOT1 (the KMT that catalyzes H3K79 methylation), all other KMTs contain the conserved SET domain (Izzo and Schneider, 2010). Lysine methylation is reversed by two classes of histone lysine demethylases. The first is the amine oxidase domain-containing enzymes, and the second class the jumonji domain-containing proteins, which are Fe(II)- and 2-oxoglutarate-dependent oxygenases. Five residues in the N-terminal tails of H3 and H4 (H3K4, H3K9, H3K27, H3K36, and H4K20) are known to be methylated and potently regulate transcription (Izzo and Schneider, 2010). Two residues in the H3 globular domain (H3K64 and H3K79), and several sites in H2A, H2B, and H1 are also methylated, although the functions of these modifications are largely unknown. H3K9, H3K27, and H4K20 methylation are associated with transcriptional silencing and heterchromatin formation/maintenance, whereas H3K4 and H3K36 methylation are associated with active transcription (Berger, 2007; Izzo and Schneider, 2010). The mechanism by which these methylation marks exert their functional effects is likely through the recruitment of different complexes of proteins with reader domains that specifically recognize these various methylation modifications.
HISTONE ACETYLATION AND METHYLATION IN DEPRESSION
Histone Acetylation
The first evidence that histone acetylation has a role in depression were observations that systemic or intracerebral administration of various HDAC inhibitors (HDACi), either alone or in combination with antidepressants, improved antidepressant responses in a variety of animal models (Covington et al, 2009, 2011b; Schroeder et al, 2007; Semba et al, 1989; Tsankova et al, 2006; Uchida et al, 2011; Weaver et al, 2004; Yamawaki et al, 2011; Zhu et al, 2009). Although one study failed to demonstrate antidepressant activity of systemically administered sodium butyrate, a non-selective HDACi, this may be explained by differences in dose as well as experimental paradigm (Gundersen and Blendy, 2009).
A study from our laboratory demonstrated that histone acetylation (H3K14ac) is transiently decreased and then persistently increased in the NAc after chronic social defeat stress, and that this is mirrored by a reduction in HDAC2 levels (Covington et al, 2009). These changes were also observed in the NAc of depression patients in postmortem examination. The observation that MS275 (a specific class I HDACi) directly infused into the NAc is robustly antidepressant initially suggests that histone acetylation has an adaptive role in stress and depression (Covington et al, 2009). A later study supported such an adaptive role of histone acetylation by demonstrating that animals overexpressing dominant-negative HDAC2 in the NAc exhibited ‘antidepressant-like’ behavior compared with control animals, whereas animals overexpressing a version of HDAC2 that strongly associates with chromatin (it lacks S-nitrosylation sites) exhibit more depression-like behavior (Uchida et al, 2011). In contrast, studies with HDAC5, a Class II HDAC, reveal that this molecule is protective and may exert resilience-like effects in the NAc. Mice susceptible to chronic social defeat stress demonstrated a decrease in HDAC5 expression whereas chronic antidepressant (imipramine) treatment increased the HDAC5 expression. In addition, mice lacking HDAC5 exhibited increased depressive-like behaviors after chronic social defeat stress compared with control animals (Renthal et al, 2007). These studies suggest that there may be at least two distinct populations of genes in the NAc mediating susceptibility to stress and depression (Figure 3). Gene targets of HDAC2 may be pro-resilient and mediate antidepressant responses, whereas those of HDAC5 may have the opposite role. Another possible explanation for HDAC2 vs HDAC5 differences could be the non-histone targets of HDAC5, as it is localized in the cytoplasm as well as the nucleus.
Epigenetic changes in the depressed brain. Summary of epigenetic changes reported to date in the brain and periphery in depressed humans or after chronic stress in animal models, as well as after antidepressant treatment (or in resilient or remitted subjects). Global as well as representative promoter-specific changes are shown. Changes marked by green arrows are thought to be adaptive/antidepressant/pro-resilient. Changes marked by red arrows are thought to be pro-depressive. Changes marked by blue arrows have as yet an unclear role in depression. Solid-line arrows represent established connections, whereas dotted-line arrows represent putative connections. In the nucleus accumbens (NAc) of a stress-susceptible mouse strain (ie, balb/c), global HDAC2 expression and HDAC2 binding on specific genes (eg, Gdnf) were increased after chronic stress (Uchida et al, 2011). These changes reversed by antidepressant treatment, while the opposite adaptive changes were observed in a more resilient stain of mice (c57b1/6) and in humans (Covington et al, 2009). On the contrary, HDAC5, a class II HDAC, were decreased in animals susceptible to stress and was increased after antidepressant treatment (Renthal et al, 2007). These results suggest that there may be two distinct groups of pro-resilient/antidepressant genes: those targeted by HDAC5 and that show decreased acetylation (and possibly decreased transcription), and those targeted by HDAC2 and that show increased acetylation (and possibly increased transcription). In addition, global levels of repressive histone methylation machinery for H3K9 (G9a, GLP, and SUV39H1), which presumably control global H3K9me2 levels, as well as H3K9me2 binding on specific genes (eg. Hras), were decreased in the NAc of susceptible animals after chronic social defeat stress, while the opposite changes were observed in resilient animals (Covington et al, 2011a). Also, histone methyltransferase (MLL1) and demethylase (LSD1) for H3K4 were decreased in susceptible animals, while they were increased in resilient animals after social defeat, with no change in global levels of H3K4 methylation (Covington et al, 2011a). The role of these changes are yet unclear. A ChIP-chip study has revealed dynamic changes in repressive histone methylation (H3K9/K27 methlation) in the upstream regulatory regions genome-wide after social defeat, with more genes showing increased methylation (eg, Ikbkb and Jundm2) and putative decreased transcription (Wilkinson et al, 2009). Many of these changes were reversed by antidepressant treatment or were not observed in resilient animals. In the hippocampus of susceptible rodents after various stress paradigms, global acetylation levels on numerous histone residues were decreased, accompanied by increased HDAC expression or activity (Covington et al, 2011b; Hollis et al, 2011; Ferland and Schrader, 2011). These changes were either not observed, returned to baseline, or changed in the opposite direction in resilient animals or after antidepressant treatment. Similar changes were observed for H3K9me3 in the hippocampus (Hunter et al, 2009). Gene-specific epigenetic changes were also observed in the hippocampus (eg, Bdnf, Nr3c1 gene cluster, and Grin2b). In the peripheral blood of depressed patients, HDAC2,4, and 5, as well as SIRT1, 2, and 6 were increased only in the depressive and not in the remissive state, while HDAC6 and 8 were increased in all patients regardless of states (Iga et al, 2007; Hobara et al, 2010; Abe et al, 2011).
Similarly, a large body of literature has suggested that histone acetylation in the hippocampus has an overall adaptive role in stress and antidepressant responses. Several studies have reported increased histone acetylation or phosphoacetylation (phosphorylation and acetylation of nearby residues in H3) in several subregions of the hippocampus after exposure to a variety of acute stressors (predator stress, social defeat stress, and forced swim stress) in both mice and rats, and that these have an adaptive role in memory formation (Bilang-Bleuel et al, 2005; Chandramohan et al, 2008; Radley et al, 2011; Reul and Chandramohan, 2007). However, opposite to what was observed in the NAc after chronic social defeat stress, there was—following a transient increase—a persistent decrease in histone acetylation (H3K14Ac) in the hippocampus (Covington et al, 2011b). These changes were reversed by imipramine, suggesting that the persistent decrease in acetylation in the hippocampus may be maladaptive, an interpretation supported by antidepressant-like responses elicited upon intra-hippocampal administration of HDACi (Covington et al, 2011b).
In a rat model of individual differences to stress, high responders (HR), which exhibit lower levels of anxiety and higher preference for sucrose solution over water when compared with low responders (LR), have higher levels of CREB-binding protein—a type of HAT, lower levels of HDAC3, and higher global levels of acetylated H3 and H2B in the hippocampus (Hollis et al, 2011). Following chronic social defeat, HR rats had a significant reduction in body mass gain, reduced sucrose preference, and decreased social interaction whereas LR rats did not demonstrate these depression-like behaviors. Correspondingly, HR rats had decreased levels of H3 and H2B acetylation in hippocampus, whereas LR rats showed increased H3 acetylation after chronic social defeat, possibly explaining their respective susceptibility and resilience to the chronic stress (Hollis et al, 2011). Curiously, levels of H4 acetylation were decreased in both groups following chronic social defeat (Hollis et al, 2011). One possible explanation for these surprising results might be the lack of specificity of the acetyl-H4 antibody used. However, a similar observation has been reported at the BDNF promoter after electroconvulsive seizures (ECS) (Tsankova et al, 2004). It was hypothesized in this latter study that the downregulation of H4 acetylation at the BDNF P2 promoter might represent the suppression of this specific promoter to allow for other promoters to override control of BDNF expression. These data thus highlight possible differential roles for H3 vs H4 acetylation, and suggest the importance of complementing studies of global histone modifications with genome-wide analyses of such changes at specific gene promoters (see below under Identification of gene targets: genome-wide and promoter-specific studies).
In another study in rats using a different depression model, chronic variable stress, decreased acetylation (H4K12Ac), and phosphoacetylation (H3K9S10) were observed in CA3 and dentate gyrus (DG) of stressed rats compared with control (Ferland and Schrader, 2011). Corresponding to the decreased acetylation, there was increased HDAC5 expression as well as HDAC Class I/II and sirtuin activities in subregions of the hippocampus (Ferland and Schrader, 2011).
The adaptive role of histone acetylation in the hippocampus in stress models has been demonstrated in several investigations. As noted above, infusion of the HDACi MS275 directly into the hippocampus was sufficient in reversing the anhedonia (measured by sucrose preference) observed after chronic social defeat stress, without affecting other measures such as social avoidance and immobility in the forced swim test (Covington et al, 2011b). This points to regional specificity in terms of regulating different aspects of the wide spectrum of behavioral disturbances observed with depression. Interestingly, MS275 infusion into the hippocampus in combination with social housing was able to reverse the social avoidance phenotype (Covington et al, 2011b). In contrast to the antidepressant-like actions of HDAC5 in the NAc, this HDAC seems to have a pro-depressive role in the hippocampus (Tsankova et al, 2006). Expression of HDAC5, but not of several other HDACs, is downregulated in the hippocampus of socially defeated mice that have been treated with imipramine (Tsankova et al, 2006). At the same time, overexpression of HDAC5 blocks the antidepressant effect of imipramine in this paradigm (Tsankova et al, 2006). Two non-exclusive explanations exist for the apparent opposite roles of HDAC5 in hippocampus vs NAc in depression models. First, HDAC5 could be part of different complexes in different brain regions and thereby target different subsets of genes that differentially contribute to depression vulnerability. Second, HDAC5 could regulate the same subsets of genes in different brain regions, with these genes mediating opposite effects on depression-related behavior. Indeed, numerous genes have been shown to exert opposite effects in depression models when acting in hippocampus vs NAc (Nestler and Carlezon, 2006). Studies that examined HDAC5 levels in the cortex of different inbred mouse strains found a negative correlation between HDAC5 levels and depressive-like behaviors, specifically, that high HDAC5 expression is associated with less depressive-like behavior (Benton et al, 2011), agreeing with observations in NAc and opposite to what is observed in hippocampus.
NAc and hippocampus are by far the most investigated regions in the study of epigenetics and depression; however, a limited amount of literature is available for other regions in the brain's cortical-striatal-limbic circuit. Following chronic social defeat stress, acetylation of H3K14 is increased in the amygdala at both 1 and 24 h after the last defeat, but returns to baseline after longer periods (Covington et al, 2011b). HDACi (MS275) infusion into the amygdala reverses social avoidance but not anhedonia, suggesting that the acetylation in the amygdala may be adaptive with respect to some aspects of depression-like behavior. In rats that have undergone acute social defeat, H3 acetylation is decreased transiently in the amygdala, although there was no effect of chronic social defeat in rats (Hollis et al, 2011). Another study examining chronic variable stress in rats found reduced HDAC5 levels in the central amygdala in male rats (Sterrenburg et al, 2011), similar to what was observed in NAc after chronic social defeat stress in mice (Renthal et al, 2007). From a limited number of studies examining the medial PFC, global levels of histone acetylation do not seem to change after acute or chronic stress in rodent models (Covington et al, 2011b; Hollis et al, 2011). More studies into these regions are necessary to expand upon these initial findings.
A large number of studies have also examined the histone acetylation machinery as potential biomarkers in peripheral blood cells of depressed human patients. One study found that HDAC2 and HDAC5 were increased in patients in their depressive but not remitted state, whereas HDAC6 and HDAC8 were decreased in patients regardless of their states, and HDAC4 was increased in the depressive state of bipolar patients (Hobara et al, 2010). Studies in mice suggest that these changes are unlikely due to antidepressant treatment (Hobara et al, 2010). The induction of HDACs 2/5 and 4 in unipolar and bipolar patients, respectively, only in the depressive state suggests that these changes may be associated with depression symptoms rather than adaptive responses, and agree with the general consensus (albeit with some exceptions) that histone acetylation in brain has a pro-adaptive role in stress and depression. Further concurring with the pro-depressive role of HDAC5 in the hippocampus, another study found that peripheral expression of HDAC5 was higher in drug-free depressive patients compared with controls, with this effect reversed after chronic antidepressant treatment (Iga et al, 2007). Although the study of sirtuins in depression is largely lacking, one study found that SIRTs 1, 2, and 6 are decreased in both MDD and bipolar patients in the depressive state but not remitted state. This finding suggests an interesting role for these class III HDACs that deserve further investigation (Abe et al, 2011).
Histone Methylation
Chronic social defeat stress has been found to decrease several histone methyltransferases (G9a, GLP, and SUV39H1), and a co-repressor (CoREST) involved in transcriptional repressive complexes, in the NAc of susceptible animals whereas these molecules are upregulated in resilient mice (Covington et al, 2011a). Specifically H3K9me2, a mark catalyzed by G9a and GLP and mainly associated with euchromatic gene repression, was also decreased in susceptible mice only (Covington et al, 2011a). Similar changes were seen in the NAc of depressed humans. Overexpression of G9a in the NAc exerted antidepressant-like behaviors, whereas knockdown of G9a exerted pro-depressant-like effects (Covington et al, 2011a). This evidence suggests that histone methylation, at least at the H3K9 residue, is pro-adaptive in response to stress and depression. Furthermore, genome-wide studies, described further below, have revealed a widespread H3K9/K27 methylation changes at the promoters of many genes in two models of depression, social defeat and social isolation, the majority of which are reversed by chronic antidepressant treatment and not observed in resilient mice (Wilkinson et al, 2009). It is curious to note that MLL and LSD1, a methyltransferase and demethylase of H3K4, respectively, are both decreased in the NAc of susceptible mice and increased in resilient animals after chronic social defeat stress (Covington et al, 2011a). These changes are associated with no global change in the methylation state of H3K4, although unpublished genome-wide data from our laboratory has demonstrated different levels of this mark at numerous specific genes in the NAc of susceptible and resilient mice.
Stress regulation of histone methylation in the hippocampus has been examined in only one study to date (Hunter et al, 2009). Acute and subchronic (7 day) stress increased global levels of H3K9me3 in the DG and CA1 regions, whereas chronic (21 days) stress decreased the same mark in these regions, and the latter effect was reversed by antidepressant treatment. H3K9me3 is typically thought to be involved in heterochromatin formation and maintenance, but has recently been shown to be regulated by several types of chronic stimuli; for example, cocaine treatment (Maze et al, 2011), as well as being involved in regulating transcription of a very small subset of genes (Jiang et al, 2010). The Hunter et al (2009) study suggests that H3K9me3 may have an adaptive role in stress models, a possibility supported by the antidepressant effects of overexpressing SETDB1 (a KMT that catalyzes H3K9me3) in broad forebrain regions (Jiang et al, 2010). Further work is needed to understand the underlying mechanisms involved, in particular, whether these adaptive effects of H3K9me3 are mediated by the repression of specific genes or, rather, of intergenic elements. One possible gene target, directly implicated in antidepressant-like responses, is H3K9me3-mediated repression of grin2b, which encodes the NR2b subunit of NMDA receptors (Jiang et al, 2010).
Acute or 7 days of stress also reduced global levels of H3K27me3 in subregions of the hippocampus, although no effects were observed after longer periods of stress, whereas acute but not prolonged stress decreased H3K9me1. The 7- and 21-day restraint stress, respectively, decreased and increased H3K4me3. More experiments are needed to understand the functional consequences of these various modes of histone methylation in hippocampus.
Identification of Gene Targets: Genome-Wide and Promoter-Specific Studies
The above sections reviewed global changes in histone acetylation and methylation machineries in various depression models, but the far more important functional question is: at what specific genes or intergenic regions does such regulation occur to control depression-related behavior? Recent work is beginning to address this key question.
BDNF is one target gene whose epigenetic regulation has been implicated in depression models. An early study demonstrated that repeated ECS, a well-known treatment of depression, induces H3 and H4 acetylation at promoter regions of the bdnf gene as well as several others (c-fos and creb) in the hippocampus, changes that correlated well with expression of these genes after ECS (Tsankova et al, 2004). In a follow-up study, it was demonstrated that chronic social defeat stress decreased total BDNF mRNA expression in hippocampus, which was reversed by chronic imipramine treatment (Tsankova et al, 2006). The decrease in BDNF mRNA was correlated with an increase in the repressive H3K27me2 mark at BDNF promoters after chronic social defeat stress, and this induction of repressive methylation was persistent despite chronic imipramine treatment (Tsankova et al, 2006). Instead, like ECS, chronic imipramine treatment caused an induction of acetylated H3 (as well as H3K4me2) on bdnf promoters that correlated with the reversal of BDNF expression (Tsankova et al, 2006).
Prenatal exposure to methylmercury causes a decrease in BDNF expression in the DG that is reversed by chronic treatment with fluoxetine (Onishchenko et al, 2008). Similar to our observations with stress, the reduction in BDNF expression after exposure to methylmercury was correlated with an increase in H3K27me3 and a reduction in H3 acetylation at bdnf promoters. This reduction in H3 acetylation, but not the induction H3K27me3, was reversed by chronic fluoxetine treatment (Onishchenko et al, 2008). It is curious to note that a single immobilization stress, which induces a transient decrease in BDNF mRNA levels in the hippocampus, also induced a transient decrease in H3 acetylation that returns to baseline by 24 h (Fuchikami et al, 2009). A study examining the induction of BDNF after chronic postnatal fluoxetine exposure in normal mice did not find corresponding changes in H3 acetylation or methylation changes, and points to the necessity for preexisting stress states for epigenetic changes associated with antidepressant treatment (Karpova et al, 2009).
Epigenetic alterations have been observed for other neurotrophic factors as well as downstream signaling proteins in regions in addition to the hippocampus. In the orbitofrontal cortex of suicide completers, it was found that expression of TrkB.T1, a truncated version of the TrkB receptor, was decreased with a corresponding increase in H3K27me3 at its gene promoter (Ernst et al, 2009). In a study of the NAc, GDNF, glial cell line-derived neurotrophic factor, was found to be pro-adaptive to stress (Uchida et al, 2011). In a more susceptible strain of mice (Balb/c), GDNF expression is decreased in NAc after chronic variable stress and reversed by chronic imipramine treatment. This was associated with decreased acetylated H3, decreased H3K4me3, and increased HDAC2/MeCP2 recruitment at the gdnf promoter, changes reversed by imipramine treatment. Acetylated H4 and H3K27me3 were not affected. In the more resilient strain of mice (c57bl/6), GDNF was increased in NAc by chronic stress, and this effect was associated with increased acetylated H3 and decreased H3K27me3. H4 acetylation was not affected and H3K4me3 was paradoxically decreased after stress. In addition, we have observed induction of RAS-MAPK-CREB signaling, potentially downstream of BDNF/TrkB, in the NAc of susceptible animals in response to chronic social defeat stress (Covington et al, 2011a). This induction of RAS expression in the NAc of susceptible animals is associated with a decrease in the repressive H3K9me2 mark, whereas the animals resilient to stress do not show changes in RAS induction, an effect accompanied by induction of H3K9me2 at the ras promoter (Covington et al, 2011a).
In a landmark series of experiments by Micheal Meaney and colleagues (Weaver et al, 2004), offspring of mothers that receive low levels of maternal care/grooming (LG) show increased stress reactivity and anxiety-related behavior in adulthood compared with those that receive high levels of maternal care/grooming (HG) (Weaver et al, 2004). LG animals show a decreased hippocampal glucocorticoid receptor mRNA expression in the hippocampus and corresponding decreased H3K9 acetylation around the GR gene promoter compared with HG animals (Weaver et al, 2004). They also show lower levels of glutamic acid decarboxylase 1 (GAD1) and corresponding decreased H3K9 binding at the gad1 promoter (Zhang et al, 2010). In a more recent study, H3K9ac difference were assessed across ∼7 Mb of chromosome 18 containing NR3C1 (GR receptor) in addition to the protocadherin gene cluster (McGowan et al, 2011). The authors found approximately equal numbers of peaks and valleys showing hyper and hypoacetylated regions in HG vs LG animals. This illustrates that epigenetic responses to maternal care is coordinated in an uneven manner in clusters across broad genomic areas, and involves changes in both genic and intergenic regions.
Despite numerous studies examining promoter-specific epigenetic changes, there are still a very limited number of genome-wide studies that have examined widespread epigenetic changes in animal models of depression. Only two genome-wide studies relating epigenetics and depression have been published thus far (Covington et al, 2009; Wilkinson et al, 2009). One study examined global gene expression profiles in the NAc after chronic social defeat stress with or without chronic administration of fluoxetine or intra-NAc MS275 (Covington et al, 2009). Chronic social defeat stress induced significant gene expression changes in a large number of genes, more of which showed a decreased rather than increased expression. This is somewhat surprising given that global levels of histone acetylation and repressive methylation in the NAc increased and decreased, respectively, after chronic social defeat stress, and necessitate the investigation of other posttranslational modifications in this stress paradigm (Covington et al, 2009, 2011a). MS275 and fluoxetine exerted similar antidepressant actions, and were able to reverse a large proportion of gene expression changes induced by chronic social defeat stress, possibly through the induction of histone acetylation. Although there was a common subset of genes similarly regulated by MS-275 and fluoxetine, a significant proportion of genes were differentially regulated across the two treatments. Genes/molecular pathways that were regulated by chronic social defeat stress but reversed after MS-275 included actin cytoskeleton reorganization machinery, transcription factors, signaling molecules, as well as a number of neurotransmitter receptors. Work is now needed to explore the functional influence of the various genes that comprise these clusters in stress models.
The second study utilized ChIP-chip to examine the distribution of repressive histone methylation—using an antibody that recognizes both H3K9me2 and H3K27me2—across the genome in the NAc in two models of depression, chronic social defeat stress, and prolonged social isolation in adulthood (Wilkinson et al, 2009). Many of the methylation changes observed in the NAc of susceptible animals after chronic social defeat stress were reversed by chronic imipramine treatment and were not observed in resilient animals. In both stress models, there were significant and dynamic changes in repressive methylation in the upstream regulatory regions genome wide, with more genes showing increased H3 methylation, agreeing with the gene expression changes after chronic stress described in the above genome-wide study (Covington et al, 2009). This is opposite to our biochemical data showing reduced global levels of H3K9me2 in the NAc after chronic social defeat stress, and no change in global levels of H3K27me2 (Covington et al, 2011a). The reason for these paradoxical findings is unknown, but likely reflects marked differences between global levels of a histone mark and changes that occur at specific genomic loci. Interestingly, a small but significant (∼20%) proportion of the genes identified in the ChIP-chip study showed similar regulation between the two depression models (Wilkinson et al, 2009). Many of the similarly regulated genes exhibited upregulation in NAc and are involved in inflammatory or cell death pathways as well as in gene regulation.
CONCLUSIONS AND FUTURE DIRECTIONS
Histone acetylation and methylation are important in numerous cellular processes, and more recently have been implicated in several psychiatric disorders including drug addiction and schizophrenia (Maze et al, 2010; Peter and Akbarian, 2011; Renthal and Nestler, 2009). It is evident from initial studies reviewed here that both of these forms of histone modification have important roles in the pathophysiology of depression. One theme that we can take away from these initial studies is that, unlike cocaine addiction where several modifications that result in a more permissive state of gene regulation also enhance behavioral responses to the drug, there are no generalizations that can be made for gene activation/repression in the pathophysiology of depression. Depending on the brain region and histone modification involved, epigenetic regulation that either enhances or represses gene transcription can be associated with depression-related behavior or its treatment. Presumably, elucidating the complex array of genes that show altered patterns of histone acetylation and methylation, as well as of several additional chromatin modifications, in several limbic brain regions will reveal how epigenetic mechanisms control lasting changes in gene expression, or regulation of intergenic regions, that influence the development and treatment of depression.
Moving forward, it is clear that much further work is necessary to fully elucidate the epigenetic basis of depression. One key question that is still not well-understood is the targeting of different epigenetic regulators to different genomic regions. This will require a large amount of genome-wide studies (next generation ChIP sequencing) in combination with various preclinical depression models and postmortem human brain tissue. Furthermore, investigation of many more posttranslational modifications, both in terms of more specific states of lysine acetylation and methylation as well as numerous novel modifications such as ADP-ribosylation, arginine methylation, novel histone variants, and so on in more regions relevant in the depression circuitry will provide crucial novel insights into the molecular pathogenesis of this syndrome. Another relatively unexplored areas of study in depression models are the ATP-dependent chromatin remodelers that physically alter the packing state of chromatin and restructure the nucleosome (including incorporating histone variants). Preliminary studies from our laboratory have already demonstrated potentially important roles of these molecules in the pathophysiology of depression (unpublished Data, Sun et al, SFN 2011). All of these studies will help us to elucidate the complex interactions among stress-regulated transcription factors, histone modifiers, and ATP-dependent remodelers and will be necessary to fully understand the global epigenetic landscape that accompanies the development of depression or its treatment. Our hope and expectation is that such studies will contribute to the development of true biomarkers of the illness and provide a unique path forward in constructing improved antidepressant treatments.
References
Abe N, Uchida S, Otsuki K, Hobara T, Yamagata H, Higuchi F et al (2011). Altered sirtuin deacetylase gene expression in patients with a mood disorder. J Psychiatr Res 45: 1106–1112.
Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C (1986). Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol 187: 591–601.
Alland L, Muhle R, Hou Jr H, Potes J, Chin L, Schreiber-Agus N et al (1997). Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387: 49–55.
Allfrey VG, Faulkner R, Mirsky AE (1964). Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA 51: 786–794. This study was one of the first to suggest that modifications on histones regulate transcription.
American Psychiatric Association (2000). Diagnostic Criteria from DSM-IV-TR. American Psychiatric Association: Washington, D.C.
Balu DT, Hodes GE, Anderson BT, Lucki I (2009). Enhanced sensitivity of the MRL/MpJ mouse to the neuroplastic and behavioral effects of chronic antidepressant treatments. Neuropsychopharmacology 34: 1764–1773.
Barrientos RM, Sprunger DB, Campeau S, Higgins EA, Watkins LR, Rudy JW et al (2003). Brain-derived neurotrophic factor mRNA downregulation produced by social isolation is blocked by intrahippocampal interleukin-1 receptor antagonist. Neuroscience 121: 847–853.
Benraiss A, Chmielnicki E, Lerner K, Roh D, Goldman SA (2001). Adenoviral brain-derived neurotrophic factor induces both neostriatal and olfactory neuronal recruitment from endogenous progenitor cells in the adult forebrain. J Neurosci 21: 6718–6731.
Benton CS, Miller BH, Skwerer S, Suzuki O, Schultz LE, Cameron MD et al (2011). Evaluating genetic markers and neurobiochemical analytes for fluoxetine response using a panel of mouse inbred strains. Psychopharmacology (Berl) 221: 297–315.
Berger SL (2007). The complex language of chromatin regulation during transcription. Nature 447: 407–412.
Berton O, McClung CA, Dileone RJ, Krishnan V, Renthal W, Russo SJ et al (2006). Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science 311: 864–868. This study was one of the first to use the social defeat stress paradigm to investigate the molecular underpinnings of depression by demostrating the pro-depressive role of BDNF in the mesolimbic dopamine pathway.
Bilang-Bleuel A, Ulbricht S, Chandramohan Y, De Carli S, Droste SK, Reul JM (2005). Psychological stress increases histone H3 phosphorylation in adult dentate gyrus granule neurons: involvement in a glucocorticoid receptor-dependent behavioural response. Eur J Neurosci 22: 1691–1700.
Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P (2008). Decoding the epigenetic language of neuronal plasticity. Neuron 60: 961–974.
Bosker FJ, Hartman CA, Nolte IM, Prins BP, Terpstra P, Posthuma D et al (2011). Poor replication of candidate genes for major depressive disorder using genome-wide association data. Mol Psychiatry 16: 516–532.
Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY et al (1996). Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84: 843–851. This study was one of the first to directly link histone acetylation with transcriptional activation by demonstrating that transcriptional co-activators are also histone acetyltransferases.
Caberlotto L, Hurd YL (2001). Neuropeptide Y Y(1) and Y(2) receptor mRNA expression in the prefrontal cortex of psychiatric subjects. Relationship of Y(2) subtype to suicidal behavior. Neuropsychopharmacology 25: 91–97.
Cary PD, Crane-Robinson C, Bradbury EM, Dixon GH (1982). Effect of acetylation on the binding of N-terminal peptides of histone H4 to DNA. Eur J Biochem 127: 137–143.
Chandramohan Y, Droste SK, Arthur JS, Reul JM (2008). The forced swimming-induced behavioural immobility response involves histone H3 phospho-acetylation and c-Fos induction in dentate gyrus granule neurons via activation of the N-methyl-D-aspartate/extracellular signal-regulated kinase/mitogen- and stress-activated kinase signalling pathway. Eur J Neurosci 27: 2701–2713.
Charney DS, Manji HK (2004). Life stress, genes, and depression: multiple pathways lead to increased risk and new opportunities for intervention. Sci STKE 2004: re5.
Choi JK, Howe LJ (2009). Histone acetylation: truth of consequences? Biochem Cell Biol 87: 139–150.
Christoffel DJ, Golden SA, Dumitriu D, Robison AJ, Janssen WG, Ahn HF et al (2011). IkappaB kinase regulates social defeat stress-induced synaptic and behavioral plasticity. J Neurosci 31: 314–321.
Cook SC, Wellman CL (2004). Chronic stress alters dendritic morphology in rat medial prefrontal cortex. J Neurobiol 60: 236–248.
Coppell AL, Pei Q, Zetterstrom TS (2003). Bi-phasic change in BDNF gene expression following antidepressant drug treatment. Neuropharmacology 44: 903–910.
Covington 3rd HE, Maze I, LaPlant QC, Vialou VF, Ohnishi YN, Berton O et al (2009). Antidepressant actions of histone deacetylase inhibitors. J Neurosci 29: 11451–11460.
Covington 3rd HE, Maze I, Sun H, Bomze HM, DeMaio KD, Wu EY et al (2011a). A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron 71: 656–670. This study demonstrated a protective role of histone methylation (G9a and H3K9me2) in the nucleus accumbens in cocaine-induced vulnerability to stress by preventing the activation of BDNF/TrkB/Ras pathway.
Covington 3rd HE, Vialou VF, LaPlant Q, Ohnishi YN, Nestler EJ (2011b). Hippocampal-dependent antidepressant-like activity of histone deacetylase inhibition. Neurosci Lett 493: 122–126.
Cryan JF, Page ME, Lucki I (2002). Noradrenergic lesions differentially alter the antidepressant-like effects of reboxetine in a modified forced swim test. Eur J Pharmacol 436: 197–205.
Drevets WC (2000). Neuroimaging studies of mood disorders. Biol Psychiatry 48: 813–829.
Drevets WC (2001). Neuroimaging and neuropathological studies of depression: implications for the cognitive-emotional features of mood disorders. Curr Opin Neurobiol 11: 240–249. A comprehensive review of neuroimaging and neuropathological findings across several brain structures in human patients suffering from primary mood disorders.
Drevets WC (2003). Neuroimaging abnormalities in the amygdala in mood disorders. Ann N Y Acad Sci 985: 420–444.
Drevets WC (2007). Orbitofrontal cortex function and structure in depression. Ann N Y Acad Sci 1121: 499–527.
Drevets WC, Savitz J, Trimble M (2008). The subgenual anterior cingulate cortex in mood disorders. CNS Spectr 13: 663–681.
Dudley KJ, Li X, Kobor MS, Kippin TE, Bredy TW (2011). Epigenetic mechanisms mediating vulnerability and resilience to psychiatric disorders. Neurosci Biobehav Rev 35: 1544–1551.
Dwivedi Y, Rizavi HS, Conley RR, Roberts RC, Tamminga CA, Pandey GN (2003). Altered gene expression of brain-derived neurotrophic factor and receptor tyrosine kinase B in postmortem brain of suicide subjects. Arch Gen Psychiatry 60: 804–815.
Epstein J, Pan H, Kocsis JH, Yang Y, Butler T, Chusid J et al (2006). Lack of ventral striatal response to positive stimuli in depressed vs normal subjects. Am J Psychiatry 163: 1784–1790.
Ernst C, Chen ES, Turecki G (2009). Histone methylation and decreased expression of TrkB.T1 in orbital frontal cortex of suicide completers. Mol Psychiatry 14: 830–832.
Fan GP (2012). DNA methylation: basic mechanisms. Neuropsychopharmacol Reviews (this issue).
Ferland CL, Schrader LA (2011). Regulation of histone acetylation in the hippocampus of chronically stressed rats: a potential role of sirtuins. Neuroscience 174: 104–114.
Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML et al (2005). Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA 102: 10604–10609.
Fu CH, Williams SC, Cleare AJ, Brammer MJ, Walsh ND, Kim J et al (2004). Attenuation of the neural response to sad faces in major depression by antidepressant treatment: a prospective, event-related functional magnetic resonance imaging study. Arch Gen Psychiatry 61: 877–889.
Fuchikami M, Morinobu S, Kurata A, Yamamoto S, Yamawaki S (2009). Single immobilization stress differentially alters the expression profile of transcripts of the brain-derived neurotrophic factor (BDNF) gene and histone acetylation at its promoters in the rat hippocampus. Int J Neuropsychopharmacol 12: 73–82.
Fumagalli F, Bedogni F, Perez J, Racagni G, Riva MA (2004). Corticostriatal brain-derived neurotrophic factor dysregulation in adult rats following prenatal stress. Eur J Neurosci 20: 1348–1354.
Gambrill AC, Barria A (2011). NMDA receptor subunit composition controls synaptogenesis and synapse stabilization. Proc Natl Acad Sci USA 108: 5855–5860.
Gao L, Cueto MA, Asselbergs F, Atadja P (2002). Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem 277: 25748–25755.
Gervasoni N, Aubry JM, Bondolfi G, Osiek C, Schwald M, Bertschy G et al (2005). Partial normalization of serum brain-derived neurotrophic factor in remitted patients after a major depressive episode. Neuropsychobiology 51: 234–238.
Giesbrecht CJ, Mackay JP, Silveira HB, Urban JH, Colmers WF (2010). Countervailing modulation of Ih by neuropeptide Y and corticotrophin-releasing factor in basolateral amygdala as a possible mechanism for their effects on stress-related behaviors. J Neurosci 30: 16970–16982.
Gold PW, Chrousos GP (2002). Organization of the stress system and its dysregulation in melancholic and atypical depression: high vs low CRH/NE states. Mol Psychiatry 7: 254–275.
Goldwater DS, Pavlides C, Hunter RG, Bloss EB, Hof PR, McEwen BS et al (2009). Structural and functional alterations to rat medial prefrontal cortex following chronic restraint stress and recovery. Neuroscience 164: 798–808.
Goshen I, Kreisel T, Ben-Menachem-Zidon O, Licht T, Weidenfeld J, Ben-Hur T et al (2008). Brain interleukin-1 mediates chronic stress-induced depression in mice via adrenocortical activation and hippocampal neurogenesis suppression. Mol Psychiatry 13: 717–728.
Grant PA, Berger SL (1999). Histone acetyltransferase complexes. Semin Cell Dev Biol 10: 169–177.
Gundersen BB, Blendy JA (2009). Effects of the histone deacetylase inhibitor sodium butyrate in models of depression and anxiety. Neuropharmacology 57: 67–74.
Hammen C (2005). Stress and depression. Annu Rev Clin Psychol 1: 293–319.
Harrison PJ (2002). The neuropathology of primary mood disorder. Brain 125: 1428–1449.
Harwood AJ (2003). Neurodevelopment and mood stabilizers. Curr Mol Med 3: 472–482.
Heilig M (2004). The NPY system in stress, anxiety and depression. Neuropeptides 38: 213–224.
Heine VM, Zareno J, Maslam S, Joels M, Lucassen PJ (2005). Chronic stress in the adult dentate gyrus reduces cell proliferation near the vasculature and VEGF and Flk-1 protein expression. Eur J Neurosci 21: 1304–1314.
Hildmann C, Riester D, Schwienhorst A (2007). Histone deacetylases—an important class of cellular regulators with a variety of functions. Appl Microbiol Biotechnol 75: 487–497.
Hobara T, Uchida S, Otsuki K, Matsubara T, Funato H, Matsuo K et al (2010). Altered gene expression of histone deacetylases in mood disorder patients. J Psychiatr Res 44: 263–270.
Hollis F, Duclot F, Gunjan A, Kabbaj M (2011). Individual differences in the effect of social defeat on anhedonia and histone acetylation in the rat hippocampus. Horm Behav 59: 331–337.
Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS (2009). Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci USA 106: 20912–20917.
Iga J, Ueno S, Yamauchi K, Numata S, Kinouchi S, Tayoshi-Shibuya S et al (2007). Altered HDAC5 and CREB mRNA expressions in the peripheral leukocytes of major depression. Prog Neuropsychopharmacol Biol Psychiatry 31: 628–632.
Izzo A, Schneider R (2010). Chatting histone modifications in mammals. Brief Funct Genomics 9: 429–443.
Jablonka E, Lamb MJ (2002). The changing concept of epigenetics. Ann N Y Acad Sci 981: 82–96.
Jiang Y, Jakovcevski M, Bharadwaj R, Connor C, Schroeder FA, Lin CL et al (2010). Setdb1 histone methyltransferase regulates mood-related behaviors and expression of the NMDA receptor subunit NR2B. J Neurosci 30: 7152–7167. This study was one of the first to implicate histone methylation, specifically Setdb1 in the forebrain, in mood-related behaviors.
Johnson JD, Campisi J, Sharkey CM, Kennedy SL, Nickerson M, Greenwood BN et al (2005). Catecholamines mediate stress-induced increases in peripheral and central inflammatory cytokines. Neuroscience 135: 1295–1307.
Karolewicz B, Szebeni K, Gilmore T, Maciag D, Stockmeier CA, Ordway GA (2009). Elevated levels of NR2A and PSD-95 in the lateral amygdala in depression. Int J Neuropsychopharmacol 12: 143–153.
Karpova NN, Lindholm J, Pruunsild P, Timmusk T, Castren E (2009). Long-lasting behavioural and molecular alterations induced by early postnatal fluoxetine exposure are restored by chronic fluoxetine treatment in adult mice. Eur Neuropsychopharmacol 19: 97–108.
Kendler KS (1983). Overview: a current perspective on twin studies of schizophrenia. Am J Psychiatry 140: 1413–1425.
Kendler KS (1998). Anna-Monika-Prize paper. Major depression and the environment: a psychiatric genetic perspective. Pharmacopsychiatry 31: 5–9.
Kessler RC (1997). The effects of stressful life events on depression. Annu Rev Psychol 48: 191–214.
Kessler RC, Berglund P, Demler O, Jin R, Koretz D, Merikangas KR et al (2003). The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R). JAMA 289: 3095–3105.
Khan SN, Khan AU (2010). Role of histone acetylation in cell physiology and diseases: an update. Clin Chim Acta 411: 1401–1411.
Koo JW, Duman RS (2008). IL-1beta is an essential mediaor of the antineurogenic and anhedonic effects of stress. Proc Natl Acad Sci USA 105: 751–756.
Koo JW, Russo SJ, Ferguson D, Nestler EJ, Duman RS (2010). Nuclear factor-kappaB is a critical mediator of stress-impaired neurogenesis and depressive behavior. Proc Natl Acad Sci USA 107: 2669–2674.
Kouzarides T (2007). Chromatin modifications and their function. Cell 128: 693–705.
Krishnan V, Han MH, Graham DL, Berton O, Renthal W, Russo SJ et al (2007). Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell 131: 391–404. This study described an extensive phenotypic characterization, and distinguishing molecular adaptations in the NAc and VTA, of mice showing susceptibility vs resilience to chronic social defeat stress.
Krishnan V, Nestler EJ (2008). The molecular neurobiology of depression. Nature 455: 894–902.
Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R (2006). Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem Biol 13: 563–567.
Lesch KP (2011). When the serotonin transporter gene meets adversity: the contribution of animal models to understanding epigenetic mechanisms in affective disorders and resilience. Curr Top Behav Neurosci 7: 251–280.
Liston C, Miller MM, Goldwater DS, Radley JJ, Rocher AB, Hof PR et al (2006). Stress-induced alterations in prefrontal cortical dendritic morphology predict selective impairments in perceptual attentional set-shifting. J Neurosci 26: 7870–7874.
Lutter LC, Judis L, Paretti RF (1992). Effects of histone acetylation on chromatin topology in vivo. Mol Cell Biol 12: 5004–5014.
Maze I, Covington 3rd HE, Dietz DM, LaPlant Q, Renthal W, Russo SJ et al (2010). Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science 327: 213–216.
Maze I, Feng J, Wilkinson MB, Sun H, Shen L, Nestler EJ (2011). Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc Natl Acad Sci USA 108: 3035–3040.
Maze I, Noh K, Allis CD (2012). Histone regulation in the CNS: basic principles of epigenetic plasticity. Neurospychopharmacol Reviews (this issue).
McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonte B, Szyf M et al (2009). Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci 12: 342–348.
McGowan PO, Suderman M, Sasaki A, Huang TC, Hallett M, Meaney MJ et al (2011). Broad epigenetic signature of maternal care in the brain of adult rats. PLoS One 6: e14739.
Monteggia LM, Luikart B, Barrot M, Theobold D, Malkovska I, Nef S et al (2007). Brain-derived neurotrophic factor conditional knockouts show gender differences in depression-related behaviors. Biol Psychiatry 61: 187–197.
Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D et al (2009). Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci 12: 1559–1566.
Murray CJ, Lopez AD (1997). Global mortality, disability, and the contribution of risk factors: Global Burden of Disease Study. Lancet 349: 1436–1442.
Mutskov V, Gerber D, Angelov D, Ausio J, Workman J, Dimitrov S (1998). Persistent interactions of core histone tails with nucleosomal DNA following acetylation and transcription factor binding. Mol Cell Biol 18: 6293–6304.
Nestler EJ, Carlezon Jr WA (2006). The mesolimbic dopamine reward circuit in depression. Biol Psychiatry 59: 1151–1159.
Nestler EJ, Gould E, Manji H, Buncan M, Duman RS, Greshenfeld HK et al (2002). Preclinical models: status of basic research in depression. Biol Psychiatry 52: 503–528.
Nestler EJ, Hyman SE (2010). Animal models of neuropsychiatric disorders. Nat Neurosci 13: 1161–1169.
Nibuya M, Morinobu S, Duman RS (1995). Regulation of BDNF and trkB mRNA in rat brain by chronic electroconvulsive seizure and antidepressant drug treatments. J Neurosci 15: 7539–7547. This study offered a thorough examination of changes in BDNF and its receptor, TrkB, in brain areas implicated in the pathophysiology of depression following stress and/or application of various antidepressant treatments.
Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y (1996). The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87: 953–959. This study was one of the first to directly link histone acetylation with transcriptional activation by demonstrating that transcriptional co-activators are also histone acetyltransferases.
Onishchenko N, Karpova N, Sabri F, Castren E, Ceccatelli S (2008). Long-lasting depression-like behavior and epigenetic changes of BDNF gene expression induced by perinatal exposure to methylmercury. J Neurochem 106: 1378–1387.
Pencea V, Bingaman KD, Wiegand SJ, Luskin MB (2001). Infusion of brain-derived neurotrophic factor into the lateral ventricle of the adult rat leads to new neurons in the parenchyma of the striatum, septum, thalamus, and hypothalamus. J Neurosci 21: 6706–6717.
Peter CJ, Akbarian S (2011). Balancing histone methylation activities in psychiatric disorders. Trends Mol Med 17: 372–379.
Radley JJ, Kabbaj M, Jacobson L, Heydendael W, Yehuda R, Herman JP (2011). Stress risk factors and stress-related pathology: neuroplasticity, epigenetics and endophenotypes. Stress 14: 481–497.
Renthal W, Maze I, Krishnan V, Covington 3rd HE, Xiao G, Kumar A et al (2007). Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron 56: 517–529.
Renthal W, Nestler EJ (2009). Chromatin regulation in drug addiction and depression. Dialogues Clin Neurosci 11: 257–268.
Reul JM, Chandramohan Y (2007). Epigenetic mechanisms in stress-related memory formation. Psychoneuroendocrinology 32 (Suppl 1): S21–S25.
Russo SJ, Wilkinson MB, Mazei-Robison MS, Dietz DM, Maze I, Krishnan V et al (2009). Nuclear factor kappa B signaling regulates neuronal morphology and cocaine reward. J Neurosci 29: 3529–3537.
Russo VEA, Martienssen RA, Riggs AD (1996). Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor Laboratory Press: Plainview, N.Y.
Schroeder FA, Lin CL, Crusio WE, Akbarian S (2007). Antidepressant-like effects of the histone deacetylase inhibitor, sodium butyrate, in the mouse. Biol Psychiatry 62: 55–64.
Schwer B, Verdin E (2008). Conserved metabolic regulatory functions of sirtuins. Cell Metab 7: 104–112.
Selvi RB, Kundu TK (2009). Reversible acetylation of chromatin: implication in regulation of gene expression, disease and therapeutics. Biotechnol J 4: 375–390.
Semba J, Kuroda Y, Takahashi R (1989). Potential antidepressant properties of subchronic GABA transaminase inhibitors in the forced swimming test in mice. Neuropsychobiology 21: 152–156.
Sheline YI (2003). Neuroimaging studies of mood disorder effects on the brain. Biol Psychiatry 54: 338–352.
Shimizu E, Hashimoto K, Okamura N, Koike K, Komatsu N, Kumakiri C et al (2003). Alterations of serum levels of brain-derived neurotrophic factor (BDNF) in depressed patients with or without antidepressants. Biol Psychiatry 54: 70–75.
Sterrenburg L, Gaszner B, Boerrigter J, Santbergen L, Bramini M, Elliott E et al (2011). Chronic stress induces sex-specific alterations in methylation and expression of corticotropin-releasing factor gene in the rat. PLoS One 6: e28128.
Strahl BD, Allis CD (2000). The language of covalent histone modifications. Nature 403: 41–45.
Sullivan PF, Neale MC, Kendler KS (2000). Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry 157: 1552–1562.
Taunton J, Hassig CA, Schreiber SL (1996). A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272: 408–411.
Thakker-Varia S, Krol JJ, Nettleton J, Bilimoria PM, Bangasser DA, Shors TJ et al (2007). The neuropeptide VGF produces antidepressant-like behavioral effects and enhances proliferation in the hippocampus. J Neurosci 27: 12156–12167.
Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ (2006). Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci 9: 519–525. This was one of the first mechanistic studies to implicate the role of epigenetic regulation, specifically histone acetylation, in animal models of depression.
Tsankova NM, Kumar A, Nestler EJ (2004). Histone modifications at gene promoter regions in rat hippocampus after acute and chronic electroconvulsive seizures. J Neurosci 24: 5603–5610.
Tse C, Sera T, Wolffe AP, Hansen JC (1998). Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol 18: 4629–4638.
Uchida S, Hara K, Kobayashi A, Otsuki K, Yamagata H, Hobara T et al (2011). Epigenetic status of Gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron 69: 359–372.
Vialou V, Feng J, Robison AJ, Nestler EJ (2012). Epigenetic mechanisms of depression and antidepressant action. Annu Rev Pharmacol Exp Ther (e-pup ahead of print).
Waddington CH (1940). Organisers & Genes. The University Press: Cambridge Eng.This was one of the first publications describing the concept of ‘epigenetics’ or how phenotypes are not entirely determined by what is coded in the DNA.
Waddington CH (1957). The Strategy of the Genes; a Discussion of Some Aspects of Theoretical Biology. Allen & Unwin: London.
Waddington CH (1968). Towards a Theoretical Biology; An IUBS Symposium. Edinburgh U.P: Edinburgh.
Wallace DL, Han MH, Graham DL, Green TA, Vialou V, Iniguez SD et al (2009). CREB regulation of nucleus accumbens excitability mediates social isolation-induced behavioral deficits. Nat Neurosci 12: 200–209.
Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S et al (2008). Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903.
Warner-Schmidt JL, Duman RS (2007). VEGF is an essential mediator of the neurogenic and behavioral actions of antidepressants. Proc Natl Acad Sci USA 104: 4647–4652.
Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR et al (2004). Epigenetic programming by maternal behavior. Nat Neurosci 7: 847–854. This study demonstrated that early life experience can have long-lasting behavioral consequences by altering the epigenetic state (DNA methylation and histone acetylation) at the GR promoter in the hippocampus.
Widdowson PS, Ordway GA, Halaris AE (1992). Reduced neuropeptide Y concentrations in suicide brain. J Neurochem 59: 73–80.
Wilkinson MB, Dias C, Magida J, Mazei-Robison M, Lobo M, Kennedy P et al (2011). A novel role of the WNT-dishevelled-GSK3beta signaling cascade in the mouse nucleus accumbens in a social defeat model of depression. J Neurosci 31: 9084–9092.
Wilkinson MB, Xiao G, Kumar A, LaPlant Q, Renthal W, Sikder D et al (2009). Imipramine treatment and resiliency exhibit similar chromatin regulation in the mouse nucleus accumbens in depression models. J Neurosci 29: 7820–7832. This study is currently the only genome-wide study to examine chromatin regulation in animal models of depression.
Willner P (2005). Chronic mild stress (CMS) revisited: consistency and behavioural-neurobiological concordance in the effects of CMS. Neuropsychobiology 52: 90–110.
Wolffe AP, Hayes JJ (1999). Chromatin disruption and modification. Nucleic Acids Res 27: 711–720.
Wu C, Morris JR (2001). Genes, genetics, and epigenetics: a correspondence. Science 293: 1103–1105.
Xu H, Steven Richardson J, Li XM (2003). Dose-related effects of chronic antidepressants on neuroprotective proteins BDNF, Bcl-2 and Cu/Zn-SOD in rat hippocampus. Neuropsychopharmacology 28: 53–62.
Yamawaki Y, Fuchikami M, Morinobu S, Segawa M, Matsumoto T, Yamawaki S (2011). Antidepressant-like effect of sodium butyrate (HDAC inhibitor) and its molecular mechanism of action in the rat hippocampus. World J Biol Psychiatry (e-pup ahead of print 3 August 2012).
Zhang DE, Nelson DA (1988). Histone acetylation in chicken erythrocytes. Rates of deacetylation in immature and mature red blood cells. Biochem J 250: 241–245.
Zhang TY, Hellstrom IC, Bagot RC, Wen X, Diorio J, Meaney MJ (2010). Maternal care and DNA methylation of a glutamic acid decarboxylase 1 promoter in rat hippocampus. J Neurosci 30: 13130–13137.
Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY (1998). Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J 17: 3155–3167.
Zhu H, Huang Q, Xu H, Niu L, Zhou JN (2009). Antidepressant-like effects of sodium butyrate in combination with estrogen in rat forced swimming test: involvement of 5-HT(1A) receptors. Behav Brain Res 196: 200–206.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Sun, H., Kennedy, P. & Nestler, E. Epigenetics of the Depressed Brain: Role of Histone Acetylation and Methylation. Neuropsychopharmacol 38, 124–137 (2013). https://doi.org/10.1038/npp.2012.73
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/npp.2012.73
Keywords
This article is cited by
-
SNORA69 is up-regulated in the lateral habenula of individuals with major depressive disorder
Scientific Reports (2024)
-
Prenatal Stress Induces Long-Term Behavioral Sex-Dependent Changes in Rats Offspring: the Role of the HPA Axis and Epigenetics
Molecular Neurobiology (2023)
-
Aberrant Histone Modification of TNFAIP3, TLR4, TNIP2, miR-146a, and miR-155 in Major Depressive Disorder
Molecular Neurobiology (2023)
-
Induced neural progenitor cells and iPS-neurons from major depressive disorder patients show altered bioenergetics and electrophysiological properties
Molecular Psychiatry (2022)
-
Three-way interaction effects of early life stress, positive parenting and FKBP5 in the development of depressive symptoms in a general population
Journal of Neural Transmission (2021)