Abstract
Emergence of suicidal ideation (TESI) during treatment with antidepressants in major depression led to a black box warning. We performed a genome-wide association study to identify genetic markers, which increase the risk for this serious side effect. TESI was evaluated in depressed in-patients (N=397) and defined by an emergence of suicidal thoughts during hospitalization without suicidal thoughts at admission using the suicide item (3) of the Hamilton Depression Rating Scale. Genotype distribution of 405.383 single-nucleotide polymorphisms (SNPs) in patients with TESI (N=32/8.1%) was compared to patients without increase in suicidal ideation (N=329/82.9%) and to a subgroup never reported suicidal ideation (N=79/19.9%). Top results were analyzed in an independent sample (N=501). None variant reached genome-wide significance, the best associated SNP was rs1630535 (p-value=1.3 × 10−7). The top 79 SNPs could be analyzed in an independent sample, and 14 variants showed nominal significant association with the same risk allele in the replication sample. A discriminant analysis classifying patients using these 79 SNPs revealed a 91% probability to classify TESI vs non-TESI cases correctly in the replication sample. Although our data need to be interpreted carefully owing to the small numbers in both cohorts, they suggest that a combination of genetic markers might indeed be used to identify patients at risk for TESI.
Similar content being viewed by others
INTRODUCTION
Major depression is a common psychiatric disease carrying a substantial loss of productivity and life quality and is associated with a significant morbidity and mortality with a suicide rate of about 2–9% (Bostwick and Pankratz, 2000; Kessler et al, 2005). Although antidepressants are the most effective treatment for depressive patients, there has been controversy if antidepressants, in particular serotonin reuptake inhibitors (SSRI), are implicated in the emergence or worsening of suicidal ideation (Masand et al, 1991; Teicher et al, 1990; Wirshing et al, 1992). Although treatment with antidepressants is associated with a significant reduction in suicides (Hall et al, 2003; Licinio and Wong, 2005; Morgan et al, 2004; Nakagawa et al, 2007; Rihmer and Akiskal, 2006) and was proven to have a suicide-preventive effect (Angst et al, 2005), there is some evidence that a subgroup of patients (4–14%) develop treatment-emergent suicidal ideation (TESI) in the first weeks following treatment initiation and dose adjustments (Jick et al, 2004; Licinio and Wong, 2005; March et al, 2007; Mulder et al, 2008; Perlis et al, 2007a; Seemuller et al, 2009). On the basis of a meta-analysis (Hammad et al, 2006), the US Food and Drug Administration (FDA), followed by regulatory bodies in Europe, issued a black box warning for a series of antidepressants, indicating that especially young patients under the age of 25 years may be at risk for this side effect (Marshall, 2004). Recent studies showed a significant decrease in diagnosis and psychopharmacological treatment of depressive episodes not only in children and adolescents, but also in adults following these warnings (Libby et al, 2007; Olfson et al, 2008; Valuck et al, 2007). Although controversially discussed (Gibbons, 2007; Jureidini, 2007; Olfson and Shaffer, 2007), there is evidence of a parallel increase of suicide rates in the United States and the Netherlands between 2003 and 2005 (Gibbons et al, 2007) for the first time in a decade. The intended effect of the FDA warning to improve supervision of patients treated with antidepressants has not yet occurred (Morrato et al, 2008). Moreover, the decline in treating major depression continued, without compensation by substitute care (Libby et al, 2009). Suicidal ideation was not only observed in patients treated with antidepressants, but also in patients treated with psychotherapy (Rucci et al, 2011). However, a recent meta-analysis of 14 911 patients suffering from a psychiatric disorder and treated with either paroxetine or placebo found a significant higher frequency of suicidal behavior in patients with major depression treated with the antidepressant (Carpenter et al, 2011). Although efforts are made to find predictors for young patients at risk (Brent et al, 2009; Posner et al, 2007), reliable predictors—including genetic markers—for adult patients are still missing.
Although large family and twin studies estimate the heritability of suicidal behavior to be in the range of 30–55% (Brent and Mann, 2005; Statham et al, 1998), no such formal evidence exists for TESI, as it is a rare and transient event not assessed by the usual genetic epidemiological methods of family and twin studies. Nonetheless, a genetic influence on this trait is likely. Up to now, there are eight reports about the association of TESI with genetic markers (for a review see, Brent et al (2010b) and Perroud (2011)). Two genome-wide association studies (GWAS) have been published to date. In the Sequenced Treatment Alternatives To Relieve Depression (STAR*D) trial, associations were found for genetic variants with the genetic loci encoding papilin (PAPLN) and the IL-28 α-receptor (IL28RA) (Laje et al, 2009). In the Genome-Based Therapeutic Drugs for Depression (GENDEP) study, a genetic marker in the vicinity of the guanine deaminase (GDA) gene was associated with emergent or worsening of suicidal ideation (Perroud et al, 2010). In candidate gene studies, associations with suicidal ideation were found with genetic markers within the genes encoding the glutamate receptors GRIK2 and GRIA3 (Laje et al, 2007), and the cyclic adenosine response-element binding protein (CREB1) (Perlis et al, 2007b) in the STAR*D cohort and the genes encoding brain-derived neurotrophic factor (BDNF), the neurotrophic tyrosine kinase receptor type 2 (NTRK2), and α-2 adrenergic receptor (ADRA2A) (Perroud et al, 2009) in GENDEP. Further candidate gene approaches revealed associations of markers within FK506-binding protein 5 (FKBP5) in the Treatment of Resistant Depression in Adolescents (TORDIA) study (Brent et al, 2010a) and in FKBP5 and ATP-binding cassette, subfamily B (MDR/TAP), member 1 (ABCB1) in a sample of depressed outpatients (Perroud et al, 2011). Previously, markers of the two glutamatergic receptor genes could be partly replicated in our Munich Antidepressant Response Signature (MARS) project (http://www.mars-depression.de), an independent sample consisting mainly of individuals of German origin (Menke et al, 2008).
To extend and complement these previous findings, we investigated associations between TESI and single-nucleotide polymorphisms (SNPs) in a GWAS in our MARS sample (Ising et al, 2009), with replication in a second independent German replication cohort.
MATERIALS AND METHODS
Discovery Sample: Patient Recruitment
We recruited 397 patients aged 18 to 75 years who were admitted to the hospital of the Max Planck Institute of Psychiatry (MPI), Munich, Germany, for the treatment of a depressive disorder presenting with a unipolar depressive episode (85.7%), bipolar disorder (12.4%, 6.1% bipolar I and 6.3% bipolar II), or other primary diagnosis with current depression (0.5% dysthymia, 0.7% adjustment disorder) between 2000 and 2007 (Hennings et al, 2009). Patients were diagnosed by psychiatrists according to the Diagnostic and Statistical Manual of Mental Disorders (DSM) IV criteria and were included in the study within the first 3 days after in-patient admission. Severity of depressive symptoms was assessed at admission by trained raters using the 21-item Hamilton Depression Rating Scale (HAM-D) (Hamilton, 1960, 1967). Patients fulfilling the criteria for at least a moderate depressive episode (HAM-D⩾14) were eligible. In addition, the revised version of the self-rating Symptom Checklist-90 (SCL 90-R) was applied at admission and the subscale scores for Somatization, Obsessive-Compulsive, Interpersonal Sensitivity, Depression, Anxiety, Hostility, Phobic Anxiety, Paranoid Ideation, and Psychoticism were calculated (Derogatis, 1983). Patients with depressive disorder due to a general medical or neurological condition were excluded (eg, Parkinson's disease, dementia, stroke, intoxication, severe infection, ischemic heart disease), as were patients with a lifetime diagnosis of drug abuse and depressive symptoms secondary to alcohol or substance abuse or dependency or patients suffering from a manic, hypomanic, or mixed episode at the time point of inclusion. We recorded ethnicity using a self-report sheet for nationality, first language, and ethnicity of the patient, and of all four grandparents. All individuals included were Caucasian, 85.1% were of German origin, and the remaining patients were of European descent (Central Europe, 6.5%; Eastern Europe, 7.8%; and Mediterranean, 0.6%). The study was approved by the local ethics committee, and written informed consent was obtained from all subjects.
The study is designed as a naturalistic pharmacogenetic study, and the MARS project (http://www.mars-depression.de) to reveal genetic biomarkers predicting clinical outcome and severe side effects (Ising et al, 2009). Data from these patients have been reported in a study describing a candidate gene approach attempting to replicate genetic associations with TESI in the STAR*D cohort (Menke et al, 2008). All patients are treated with antidepressants according to doctor's choice. Concomitant psychotropic medication with mood stabilizers, neuroleptics, benzodiazepines, and hypnotics was allowed. For all patients, plasma concentrations of antidepressants were monitored to assure clinically effective drug levels.
Psychopathology and Phenotype Definition
HAM-D ratings were performed within 5 days of admission and then weekly until discharge. The severity of suicidal ideation was rated by item 3 ‘suicide’ of the HAM-D, with 0 ‘absent’; 1 ‘subject feels life is not worth living’; 2 ‘wishes he/she were dead or any thoughts of possible death to self’; 3 ‘suicidal ideas or gestures’; and 4 ‘attempts at suicide’. TESI was defined as an emergence of suicidal thoughts in patients without suicidal ideation (item 3 of the HAM-D=0) at hospital admission (n=32). The non-TESI comparison groups were (1) all individuals without increase in suicidality independent of baseline suicidality (no increase of the HAM-D item 3 over time; n=329)—broader non-TESI group and (2) a subgroup of patients rating zero on item 3 throughout treatment (item 3 of the HAM-D=0 at all visits; n=79)—narrow non-TESI group. In all, 36 subjects did report suicidal ideation at admission and showed an increase in the HAM-D item 3 over the 12 weeks, so that they were not included in the initial analysis. In the STAR*D trial, the suicide item (item number 12) of the Quick Inventory of Depression Symptomatology-Self Report (QIDS-SR) was used to define TESI. Both scales should identify similar patients as the QIDS-SR has been shown to correlate well with the HAM-D score (Rush et al, 2003, 2005, 2006; Trivedi et al, 2004). The observation period for TESI was the first 12 weeks following in-patient admission, congruent with the observation period used in Laje et al. (2007). Genetic associations of GRIK2 and GRIA3 have already been reported for this sample in Menke et al. (2008).
Replication Sample
The German replication sample consisted of 501 Caucasian in-patients from the psychiatric hospital of the University of Münster and from a second wave of patient recruitment in the MPI of Psychiatry, Munich (recruited in 2007–2010). In total, 25 subjects did report suicidal ideation at admission and showed an increase in the HAM-D item 3 over the treatment period, so that they were not included in the analysis. Gender distribution (p>0.2) and age (p>0.9) did not differ between the discovery and replication samples. Overall, 85% of these patients suffered from major depression, whereas 15% were patients with bipolar disorder with a current depressive episode. Psychiatrists ascertained DSM IV diagnosis. Patients were rated weekly from admission to discharge (Munich) or until week 6 (Münster) using the 21-item HAM-D rating scale. TESI was evaluated until week 6, as TESI emerged in most cases within the first 2 weeks in the discovery sample. Ethnicity was recorded using the same self-report questionnaire as in the MARS study. All patients were Caucasian and 90.7% were of German origin; the remaining patients were of European descent (Central Europe: 3.9%; Eastern Europe: 5.3%; Mediterranean: 0.1%). Same inclusion/exclusion criteria were applied as in the MARS sample, and outcome under antidepressant treatment was evaluated accordingly.
DNA Preparation
DNA was isolated from EDTA anticoagulated venous blood samples using standardized protocols (Domschke et al, 2008; Ising et al, 2009).
SNP Genotyping
Genotyping in the discovery sample was performed on Illumina Human-1 Genotyping 100k BeadChips (109 000 SNPs selected with an exon-centric focus) and Illumina HumanHap300-Duo Genotyping BeadChips (318 000 tag SNPs) (Illumina, San Diego, CA, USA) according to the manufacturer's standard protocols. The average call rate exceeded 99%, with samples below 98% being either retyped or excluded from the study. The reproducibility for samples genotyped twice was 99.99% or better.
Neither the genomic control method (Devlin et al, 2001) applied on a genome-wide level nor the EIGENSTRAT analysis (Price et al, 2006) gave any indication for population stratification, see also Ising et al (2009); however, a multidimensional scaling plot generated from the GWAS data using PLINK (http://www.pngu.mgh.harvard.edu/~purcell/plink/) indicated that two individuals in the non-TESI group A (broad definition), of which one also belonged to the non-TESI group B (narrow definition), were more than 6 SDs from the bulk of the patients, see Supplementary Figure 1. We therefore re-ran the top 100 associations excluding these two individuals.
For the replication study, SNPs were genotyped on a MALDI-TOF mass spectrometer (MassArray system) employing the Spectrodesigner software (Sequenom, San Diego, CA, USA) for primer selection and multiplexing (I-plex), and the homogeneous mass-extension process for producing primer extension products (Tang et al, 1999). All primer sequences and assay protocols are available upon request.
Statistical Analysis of Genetic Associations
Exact tests on Hardy–Weinberg equilibrium (HWE) were performed for all SNPs (Wigginton et al, 2005). SNPs with a minor allele frequency below 2.5%, with a call rate of <98%, or displaying HWE deviation at an error level below 10−5 were excluded from the analysis. Final analysis was carried out in 371.335 SNPs. Both allelic and genotypic association tests were applied. To avoid false-positive associations due to small sample sizes, we used permutation-based p-value estimates (100 000 permutations, 100 000 000 permutations for the top 4 SNPs) in addition to asymptotic p-values. If not otherwise specified, permutation-based p-values are reported. Genetic association was tested using the WG-permer, a C++-based statistical program for rapid permutation (http://www.wg-permer.org). Phenotypic analyses were performed using SPSS version 16.0.
Selection of SNPs for Replication and Analysis in a Second Sample
A Fisher Product Method (FM) was applied over allelic and genotypic tests. Only SNPs that had an FM p-value <0.001 for TESI in comparison against both non-TESI groups A and B were considered for replication. From 100 SNPs that met the replication criteria, 79 SNPs could be successfully genotyped in the replication sample (21 SNPs were dropped owing to a call rate smaller than 98%). Permutation-based allelic and genotypic association tests were applied to test for differences in the genotype and allele distribution between the TESI and the broader non-TESI group (no increase in suicidal ideation). To correct for multiple testing, the permutation-based minimum p-method proposed by Westfall and Young was applied (Westfall, 1993), correcting for 79 SNPs and two genetic models (allelic and genotypic).
RESULTS
Sociodemographic and Clinical Differences between Groups
The comparisons of disease-related and sociodemographic variables between TESI-positive and -negative patients using both the broader (comparison group A) and more restricted definition (comparison group B) are listed in Table 1. There were no significant differences in sociodemographic factors among these groups. Except for the age at onset, there were no differences in disease-related variables at admission between the three groups, especially no differences in history of suicide attempts or severity of depression in the discovery sample. However, individuals developing TESI displayed a significantly worse response to treatment with antidepressants as compared with subjects without TESI, as measured with the HAM-D throughout a 12-week period (see Figure 1). This worse response and remission rate of the TESI-positive group was also detected in the independent replication sample; however, in this sample there were also differences in the severity of depression at admission (see Table 2).
Hamilton Depression Rating Scale (HAM-D) scores over the first 12 weeks of hospitalization plotted against treatment-emergent suicidal ideation (TESI)-positive and -negative patients. A repeated measures analysis of variance (ANOVA) showed a significant effect of interaction between TESI and non-TESI group B (F3.71,92=3.06; p=0.02) and HAM-D score. There was a significant main effect of TESI status (F1,92=18.7; p=0.000038), and no other significant interaction or main effects were observed for sex or age. Comparing TESI with non-TESI group A, a repeated measures ANOVA revealed a significant interaction with HAM-D score (F4.6,333=6.98; p=0.000004). For comparison with non-TESI group A, there was a significant interaction effect between TESI status and HAM-D score (F4.6,333=6.98; p=0.000004) and age (F4.6,333=5.4; p=0.0001). We observed a significant main effect of TESI status (F1,333=9.9; p=0.002).
TESI occurred in 8.1% of all patients, and within these, 59.3% experienced TESI within the first 2 weeks after hospital admission. Current medication at the onset of suicidal ideation was SSRIs in 37.5%, venlafaxine or duloxetine in 25%, mirtazapine in 28%, tricyclic antidepressants in 34.3%, and reboxetine in 12.5%. Antidepressant monotherapy was administered to 62.5% of the patients, whereas 37.5% received combination therapy. There were no significant differences in psychotropic medication among the TESI and non-TESI groups, except for the increased administration of benzodiazepines in the TESI group (Table 1). Benzodiazepines were prescribed for agitation, suicidal ideation, anxiety, and insomnia.
Genome-Wide Association Study
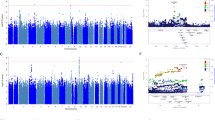
Comparing genotype and allele frequencies of all SNPs between TESI groups, we found a series of significant associations, but none of them with genome-wide significance, that is, a p-value <5.0 × 10−8 (Dudbridge and Gusnanto, 2008). The best associated SNP was rs1630535, which showed a p-value of 1.3 × 10−7, allelic OR 10.535, CI 4.2–26.4 compared with narrow non-TESI group, and a p-value of 2.45 × 10−7, OR 4.16, CI 2.3–7.4 compared with the broader non-TESI group. Rs1630535 is intergenic, the nearest gene is annexin A2 (129 kb distal of the variant), which is involved in cellular growth and signal transduction. Using the FM over allelic and genotypic models, we selected 100 SNPs, which were significantly associated with TESI with an empiric p-value <0.001 when combining the p-values for the association of TESI compared with the no increase in TESI and the never TESI group (see Supplementary Table 1). When restricting the analysis to individuals with unipolar depression (N=29/280), all 100 selected SNPs still showed association with TESI with p<0.05. After excluding the two patients with a likely different ethnic background based on multidimensional plots, also all 100 SNP associations remained nominally significant.
Replication in Independent Sample
After quality control, 79 of 100 SNPs could be analyzed in a second independent sample. Although none of the SNPs achieved significance after correction for multiple testing, 14 SNPs showed nominally significant associations (allelic or genotypic tests, p<0.05), with TESI in comparison with the broader non-TESI group and displayed the same risk alleles (see Table 3). Having 14 of 79 SNPs that show replication in the same direction with a p<0.05 is more than expected by chance, which would be 1.92 SNPs when considering allelic and genotypic tests and no linkage disequilibrium (LD) between SNPs. In the set of 14 SNPs, 6 are in high LD. However, even with a conservative estimate that only nine independent SNPs replicate in the same direction, this is still four times the number of associations than expected by chance. In the discovery sample, these 14 SNPs showed no associations with previous suicide attempts, unipolar depression, psychotic features, or response parameters like response and remission at 5 weeks. Also, there was no association with psychopathology measures from the self-assessed SCL 90-R.
Discriminant Analysis
Discriminant analysis of the 79 genotyped SNPs within the replication sample (independent variables) and TESI (group variable) showed the 79 SNPs to have highly significant discriminant power (Wilk's λ=0.454; χ2=111.039; d.f.=78; p=0.003) and standardized canonical discriminant function analysis showed significant contributions of the 79 SNPs (canonical correlation coefficient: 0.739) in the replication sample using the broad non-TESI definition. Discriminant analysis using the narrow non-TESI definition in the replication sample showed: Wilk's λ=0.094; χ2=85.276; d.f.=78; p=0.018; and canonical correlation coefficient: 0.952. Overall, the discriminant analysis using this subset of 79 SNPs revealed a 91% probability to classify TESI vs broad non-TESI cases correctly in the replication sample, and 73% when using the narrow non-TESI group. Specificity and sensitivity in the replication sample were 96%/36% using the broad non-TESI group and 79%/52% using the narrow non-TESI group. Therefore, we observed a high negative predictive value with 94% (patients who did not suffer from TESI), but only a modest positive predictive value with 48% (patients who did develop TESI).
Replication of STAR*D SNPs
The GWAS of TESI by Laje et al. (2009) found two SNPs withstanding correction for multiple testing, rs11628713 (located within PAPLN gene) and rs10903034 (located within IL28RA). In our discovery sample, rs11628713 had an allelic p-value of 0.85 and 1, against narrow and broader non-TESI group, respectively. For rs10903034, allelic p-values were 0.3 against narrow and 0.6 against broader non-TESI group. For both SNPs genotypic p-values were >0.5. Beyond the two reported SNPs, we analyzed 17 SNPs within IL28RA and 15 SNPs within PAPLN. Two SNPs within IL28RA showed barely a nominal significance, rs11587500 with genotypic p=0.02 and allelic p=0.66 against narrow non-TESI group and genotypic p=0.07 and allelic p=0.89 against broader non-TESI group. The distinct discrepancies between genotypic and allelic tests may be due to a heterozygote advantage/disadvantage model. The second SNP was rs4649203 with genotypic p=0.09 and allelic p=0.06 against narrow non-TESI group and genotypic p=0.07 and allelic p=0.06 against broader non-TESI group. Both SNPs were not in LD with the reported STAR*D SNPs. Perlis et al (2007b) investigated five SNPs located in the cyclic adenosine monophosphate response element binding protein (CREB) in the STAR*D study and found two SNPs associated with TESI in male depressed outpatients. We analyzed eight SNPs within CREB, but owing to our sample size, we did not separate by gender. None of these SNPs were significantly associated with TESI in our discovery sample.
Replication of GENDEP SNPs
In the GWAS approach, the best associated SNP rs11143230 was located in the vicinity of GDA. We analyzed 68 SNPs (rs11143230 was not genotyped) within GDA in our discovery sample. One SNP, rs10869127, was significantly associated with TESI (tested against the non-TESI broad group), with nominal allelic p=0.037 and genotypic p=0.102. In the candidate gene approach, nine candidate genes were analyzed: HTR1A, HTR2A, TPH1, TPH2, SLC6A4, ADRA2A, SLC6A2, BDNF, and NTRK2. SNPs within BDNF, NTRK2, and ADRA2A were significantly associated in GENDEP. In our discovery sample, we investigated 165 SNPs within the reported candidate genes. Only one SNP achieved nominal significance: rs10042486 (HTR1A) with genotypic p=0.047 and allelic p=0.3. When applying the GENDEP phenotype definition, that is, the increase of suicidal ideation (n=68/17%), none of the 165 SNPs showed nominal significant associations.
Replication of TORDIA and Outpatient Sample SNPs
Two additional candidate gene approaches investigated suicidal ideation and revealed markers within FKBP5 (rs1360780, rs3800373 TORDIA; rs1360780 also the outpatient sample reported by Perroud (2011)) and within ABCB1 (2677G>T in the outpatient sample) to be associated with TESI. We investigated 28 SNPs within FKBP5 and 96 SNPs within ABCB1 in our discovery sample. Six SNPs (non-TESI narrow group: rs10246878, rs1202171, rs1202172, rs1202179, rs1202186, rs2214102, with genotypic p<0.05) and five SNPs (non-TESI broad group: rs10246878, rs1045642, rs2214102, rs2214102, rs2235046, rs2235048, with allelic/genotypic p<0.05) within ABCB1 were nominally associated with TESI. SNP rs7757037 within FKBP5 was significantly associated with TESI in the non-TESI broader group (genotypic p=0.0023) and in the non-TESI narrow group (genotypic p=0.0039). SNP rs1360780 was not significantly associated (allelic p=0.4; genotypic p=0.53, rs3800373 was not genotyped).
DISCUSSION
In this GWAS for TESI, we found a subset of 14 SNPs associated with TESI with supportive genetic evidence in a second independent sample. These 14 SNPs were part of 79 variants associated with a p<0.001 in the discovery sample and analyzed in the replication sample. This is more than would be expected by chance (even when considering that 6 of 14 SNPs are in high LD), as one would expect to only see 1.975 SNPs associated with p<0.05 in the same direction by chance. Although the results have to be interpreted cautiously and have to await further replication, we report preliminary evidence that the 79 SNPs identified in our discovery set allow to classify TESI patients with a probability of 90% in the independent replication sample. While we observed a high negative predictive value of 94% and the modest positive predictive value of 48%, identifying patients who will develop TESI may limit the clinical usefulness of this set and indicates that more studies in larger samples will be necessary for genetic markers that can be used in clinical practice.
We also investigated whether the findings reported in the GWAS of TESI in the STAR*D trial (Laje et al, 2009) and the previously reported associations within the CREB gene (Perlis et al, 2007a) would replicate in our sample. None of the SNPs with reported TESI association in the STAR*D cohort achieved nominal significance in our discovery sample. For a gene-wise replication approach, we additionally analyzed 17 IL28RA and 15 PAPLN SNPs, as well as 8 CREB SNPs, and found only one weak association for one IL28RA SNP, which is not in LD with the SNPs associated in the STAR*D sample. However, the CREB association findings were generated with a slightly different phenotype definition, individuals developing TESI could have a score of 0 or 1 at the initial HAM-D suicide item, while the GWAS, as well as our study demanded an initial score of 0. In addition, we analyzed 165 SNPs within the candidate genes reported in the GENDEP trial. None of the SNPs located in BDNF or NTRK2 were significantly associated in our discovery sample. However, we found one SNP within GDA, which was associated in the GENDEP GWAS approach, significantly associated with TESI. Moreover, we found further evidence for FKBP5 and ABCB1 to be implicated in TESI, which were previously reported in two other candidate gene approaches on emerging or worsening of suicidal ideation (Brent et al, 2010a; Perroud, 2011).
GENDEP investigated not only the worsening of suicidal ideation, but also the emergence that increased the proportion of affected patients to 32%. Negative replication may thus be the result of different phenotype definitions. This is supported by results from a secondary analysis (data not shown) in which the previously excluded patients, which presented suicidal ideation at admission and then exhibited worsening of suicidal ideation (n=36), were included increasing the TESI-positive participants to n=68. However, none of the p-values of the 14 replicate SNPs did not get stronger, suggesting that emergent and worsening suicidal ideation may not share the same genetic risk factors. In contrast, the STAR*D sample and our sample only include patients with emergent suicidal ideation and have a very low number of individuals affected by TESI, and therefore have a very high potential for false positive as well as negative associations. Further studies in larger samples will be needed to replicate or falsify any given locus. In addition, there are some major differences between the two samples that could also explain incongruent results. These range from a different ethnic composition, differences in disease-related variables such as rate of chronic depression or inclusion of patients with bipolar disorder over differences in rating scales to differences in study design—monotherapy with citalopram in outpatients in STAR*D against a psychopharmacological combination therapy with diverse antidepressants and severely depressed in-patients in MARS.
Most of the TESI-related studies (Laje et al, 2007; Perlis et al, 2007b), including ours, have relied on a single rating scale item to capture suicidal ideation. In these studies, the feeling of hopelessness is already rated as suicidal ideation, but may in fact not necessarily represent a wish of death. Although this may limit the diagnostic specificity, the rate and the occurrence time of the TESI-affected individuals was similar in various studies, with 6.9% in STAR*D, 11.4% in an outpatient sample of New Zealand (Mulder et al, 2008), 7.8% in a sample of elderly depressed patients (Szanto et al, 2007), 8.1% in our discovery sample and 8.4% in the replication sample. Furthermore, suicidal ideation appeared within the first weeks of treatment initiation or dose increase and would thus be consistent with treatment emergence (Juurlink et al, 2006; Laje et al, 2007; Mulder et al, 2008).
Meta-analysis of antidepressant trials suggested that adolescents and young adults up to the age of 25 years have the greatest risk of developing suicidal ideation during treatment (Leon, 2007). However, in our study, as well as in STAR*D (Laje et al, 2007), there was no age difference between TESI-positive and -negative patients. In addition, common risk factors for suicide attempts or completed suicides such as a history of attempted suicide, duration of the illness, or the current depressive episode, employment, and relationship status (Mann, 2005) were not associated with TESI. This again is consistent with observations in STAR*D (Laje et al, 2007; Zisook et al, 2009). In our study, we only applied the structured interview data for Axis II comorbidity, so that influence of comorbid personality disorder on TESI could not be analyzed. However, we can report the self-assessed features of the SCL 90 for the discovery sample. Only the subscales representing depression and psychoticism were significantly different between TESI-positive and non-TESI patients. Using these scores as covariates did not change the association of the top 100 SNPs with TESI in the discovery sample (data not shown).
There is an ongoing debate about the TESI phenotype. The appearance of suicidal ideation during treatment with antidepressants may just be a marker for therapy non-response and not an independent phenotype as the main difference between the affected and non-affected patients across all studies are reduced response and remission rates (Laje et al, 2007; Szanto et al, 2007). If so, genetic predictors of suicidal-adverse events may also predict poor response to treatment (Brent et al, 2009). However, none of the 14 SNPs predicting TESI in both samples were associated with response parameters. In addition, none of these SNPs were associated with unipolar depression, previous suicide attempts, or psychotic features. We also analyzed the 14 SNPs in four patients who committed suicide during treatment or shortly after discharge. These patients carried the risk allele in an average of 64% (57–77%) SNPs. Owing to suicidal ideation at admission, they were not classified as TESI positive.
None of the genes harboring the 14 SNPs have yet been reported to be associated with suicidality in general. However, the genetic loci containing the genes RHEB, TMEM138, and CYBASC3 that are associated with TESI have shown linkage with bipolar disorder (Cassidy et al, 2007; Elashoff et al, 2007; Pato et al, 2004). PIK3C3 is considered as a candidate gene for schizophrenia (Carter, 2009; Tang et al, 2008) and was previously associated with bipolar disorder and neurodegeneration (Stopkova et al, 2004; Wang et al, 2011). Hence, nine variants were located in or nearby genes, which were previously linked to bipolar disorder and one variant in a gene also associated with schizophrenia and neurodegeneration. However, without any functional evidence that these variants do indeed alter the function of these genes, one has to be very cautious with such extrapolations. We consider the 14 variants as biomarkers for TESI, which do not necessarily have to be directly related to pathophysiology.
An important limitation of our study is the size of both samples. The number of TESI-affected individuals is low in both the German discovery and replication samples as TESI is an uncommon phenotype (Licinio and Wong, 2005). Even the largest pharmacogenetics of antidepressant study to date, the STAR*D trial, with over 1900 patients included only yields 91 patients affected with TESI. Therefore, validation of association results with TESI is currently very difficult and will need national and international consortia to generate samples of sufficient size. Another limitation of all current TESI genetic studies is the lack of a placebo comparator, so that the emergence of suicidal ideation cannot specifically be attributed to the pharmacological treatment, but could also reflect a general worsening of the depressive symptoms. In our study, the limited sample size does not allow one to investigate whether there are differential associations of these SNPs with TESI in patients with unipolar vs bipolar disorder, but all associations remain significant when restricting the analysis to unipolar depression.
Even though the genetics of TESI are limited by the small number of affected cases, even in large pharmacogenetic studies, this study—as well as previous reports—suggests that genetic markers may be used as tools to identify patients at risk for this serious side effect. This would allow one to provide patients at risk with closer monitoring following antidepressant treatment initiation.
References
Angst J, Angst F, Gerber-Werder R, Gamma A (2005). Suicide in 406 mood-disorder patients with and without long-term medication: a 40 to 44 years’′ follow-up. Arch Suicide Res 9: 279–300.
Bostwick JM, Pankratz VS (2000). Affective disorders and suicide risk: a reexamination. Am J Psychiatry 157: 1925–1932.
Brent D, Melhem N, Ferrell R, Emslie G, Wagner KD, Ryan N et al (2010a). Association of FKBP5 polymorphisms with suicidal events in the Treatment of Resistant Depression in Adolescents (TORDIA) study. Am J Psychiatry 167: 190–197.
Brent D, Melhem N, Turecki G (2010b). Pharmacogenomics of suicidal events. Pharmacogenomics 11: 793–807.
Brent DA, Emslie GJ, Clarke GN, Asarnow J, Spirito A, Ritz L et al (2009). Predictors of spontaneous and systematically assessed suicidal adverse events in the treatment of SSRI-resistant depression in adolescents (TORDIA) study. Am J Psychiatry 166: 418–426.
Brent DA, Mann JJ (2005). Family genetic studies, suicide, and suicidal behavior. Am J Med Genet C 133: 13–24.
Carpenter DJ, Fong R, Kraus JE, Davies JT, Moore C, Thase ME (2011). Meta-analysis of efficacy and treatment-emergent suicidality in adults by psychiatric indication and age subgroup following initiation of paroxetine therapy: a complete set of randomized placebo-controlled trials. J Clin Psychiatry (in press).
Carter CJ (2009). Schizophrenia susceptibility genes directly implicated in the life cycles of pathogens: cytomegalovirus, influenza, herpes simplex, rubella, and Toxoplasma gondii. Schizophr Bull 35: 1163–1182.
Cassidy F, Zhao C, Badger J, Claffey E, Dobrin S, Roche S et al (2007). Genome-wide scan of bipolar disorder and investigation of population stratification effects on linkage: support for susceptibility loci at 4q21, 7q36, 9p21, 12q24, 14q24, and 16p13. Am J Med Genet B 144B: 791–801.
Derogatis LR (1983). Misuse of the symptom checklist 90. Arch Gen Psychiatry 40: 1152–1153.
Devlin B, Roeder K, Wasserman L (2001). Genomic control, a new approach to genetic-based association studies. Theor Popul Biol 60: 155–166.
Domschke K, Dannlowski U, Ohrmann P, Lawford B, Bauer J, Kugel H et al (2008). Cannabinoid receptor 1 (CNR1) gene: impact on antidepressant treatment response and emotion processing in major depression. Eur Neuropsychopharmacol 18: 751–759.
Dudbridge F, Gusnanto A (2008). Estimation of significance thresholds for genomewide association scans. Genet Epidemiol 32: 227–234.
Elashoff M, Higgs BW, Yolken RH, Knable MB, Weis S, Webster MJ et al (2007). Meta-analysis of 12 genomic studies in bipolar disorder. J Mol Neurosci 31: 221–243.
Gibbons RD (2007). Dr. Gibbons Replies. Am J Psychiatry 164: 1908–1910.
Gibbons RD, Brown CH, Hur K, Marcus SM, Bhaumik DK, Erkens JA et al (2007). Early evidence on the effects of regulators’ suicidality warnings on SSRI prescriptions and suicide in children and adolescents. Am J Psychiatry 164: 1356–1363.
Hall WD, Mant A, Mitchell PB, Rendle VA, Hickie IB, McManus P (2003). Association between antidepressant prescribing and suicide in Australia, 1991–2000: trend analysis. BMJ 326: 1008.
Hamilton M (1960). A rating scale for depression. J Neurol Neurosurg Psychiatry 23: 56–62.
Hamilton M (1967). Development of a rating scale for primary depressive illness. Br J Soc Clin Psychol 6: 278–296.
Hammad TA, Laughren T, Racoosin J (2006). Suicidality in Pediatric Patients Treated With Antidepressant Drugs. Arch Gen Psychiatry 63: 332–339.
Hennings JM, Owashi T, Binder EB, Horstmann S, Menke A, Kloiber S et al (2009). Clinical characteristics and treatment outcome in a representative sample of depressed inpatients—findings from the Munich Antidepressant Response Signature (MARS) project. J Psychiatr Res 43: 215–229.
Ising M, Lucae S, Binder EB, Bettecken T, Uhr M, Ripke S et al (2009). A genomewide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Arch Gen Psychiatry 66: 966–975.
Jick H, Kaye JA, Jick SS (2004). Antidepressants and the risk of suicidal behaviors. Jama 292: 338–343.
Jureidini J (2007). The Black Box Warning: decreased prescriptions and increased youth suicide? Am J Psychiatry 164: 1907.
Juurlink DN, Mamdani MM, Kopp A, Redelmeier DA (2006). The risk of suicide with selective serotonin reuptake inhibitors in the elderly. Am J Psychiatry 163: 813–821.
Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE (2005). Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry 62: 593–602.
Laje G, Allen AS, Akula N, Manji H, John Rush A, McMahon FJ (2009). Genome-wide association study of suicidal ideation emerging during citalopram treatment of depressed outpatients. Pharmacogenet Genomics 19: 666–674.
Laje G, Paddock S, Manji H, Rush AJ, Wilson AF, Charney D et al (2007). Genetic markers of suicidal ideation emerging during citalopram treatment of major depression. Am J Psychiatry 164: 1530–1538.
Leon AC (2007). The revised warning for antidepressants and suicidality: unveiling the black box of statistical analyses. Am J Psychiatry 164: 1786–1789.
Libby AM, Brent DA, Morrato EH, Orton HD, Allen R, Valuck RJ (2007). Decline in treatment of pediatric depression after FDA advisory on risk of suicidality with SSRIs. Am J Psychiatry 164: 884–891.
Libby AM, Orton HD, Valuck RJ (2009). Persisting decline in depression treatment after FDA warnings. Arch Gen Psychiatry 66: 633–639.
Licinio J, Wong ML (2005). Depression, antidepressants and suicidality: a critical appraisal. Nat Rev Drug Discov 4: 165–171.
Mann JJ (2005). The medical management of depression. N Engl J Med 353: 1819–1834.
March JS, Silva S, Petrycki S, Curry J, Wells K, Fairbank J et al (2007). The Treatment for Adolescents With Depression Study (TADS): long-term effectiveness and safety outcomes. Arch Gen Psychiatry 64: 1132–1143.
Marshall E (2004). Antidepressants and children: buried data can be hazardous to a company's health. Science 304: 1576–1577.
Masand P, Gupta S, Dewan M (1991). Suicidal ideation related to fluoxetine treatment. N Engl J Med 324: 420.
Menke A, Lucae S, Kloiber S, Horstmann S, Bettecken T, Uhr M et al (2008). Genetic markers within glutamate receptors associated with antidepressant treatment-emergent suicidal ideation. Am J Psychiatry 165: 917–918.
Morgan OW, Griffiths C, Majeed A (2004). Association between mortality from suicide in England and antidepressant prescribing: an ecological study. BMC Public Health 4: 63.
Morrato EH, Libby AM, Orton HD, Degruy III FV, Brent DA, Allen R et al (2008). Frequency of provider contact after FDA advisory on risk of pediatric suicidality with SSRIs. Am J Psychiatry 165: 42–50.
Mulder RT, Joyce PR, Frampton CM, Luty SE (2008). Antidepressant treatment is associated with a reduction in suicidal ideation and suicide attempts. Acta Psychiatr Scand 118: 116–122.
Nakagawa A, Grunebaum MF, Ellis SP, Oquendo MA, Kashima H, Gibbons RD et al (2007). Association of suicide and antidepressant prescription rates in Japan, 1999–2003. J Clin Psychiatry 68: 908–916.
Olfson M, Marcus SC, Druss BG (2008). Effects of Food and Drug Administration warnings on antidepressant use in a national sample. Arch Gen Psychiatry 65: 94–101.
Olfson M, Shaffer D (2007). SSRI prescriptions and the rate of suicide. Am J Psychiatry 164: 1907–1908.
Pato CN, Pato MT, Kirby A, Petryshen TL, Medeiros H, Carvalho C et al (2004). Genome-wide scan in Portuguese Island families implicates multiple loci in bipolar disorder: fine mapping adds support on chromosomes 6 and 11. Am J Med Genet B 127B: 30–34.
Perlis RH, Beasley Jr CM, Wines Jr JD, Tamura RN, Cusin C, Shear D et al (2007a). Treatment-associated suicidal ideation and adverse effects in an open, multicenter trial of fluoxetine for major depressive episodes. Psychother Psychosom 76: 40–46.
Perlis RH, Purcell S, Fava M, Fagerness J, Rush AJ, Trivedi MH et al (2007b). Association between treatment-emergent suicidal ideation with citalopram and polymorphisms near cyclic adenosine monophosphate response element binding protein in the STAR*D study. Arch Gen Psychiatry 64: 689–697.
Perroud N (2011). Suicidal ideation during antidepressant treatment: do genetic predictors exist? CNS Drugs 25: 459–471.
Perroud N, Aitchison KJ, Uher R, Smith R, Huezo-Diaz P, Marusic A et al (2009). Genetic predictors of increase in suicidal ideation during antidepressant treatment in the GENDEP project. Neuropsychopharmacology 34: 2517–2528.
Perroud N, Bondolfi G, Uher R, Gex-Fabry M, Aubry JM, Bertschy G et al (2011). Clinical and genetic correlates of suicidal ideation during antidepressant treatment in a depressed outpatient sample. Pharmacogenomics 12: 365–377.
Perroud N, Uher R, Ng MY, Guipponi M, Hauser J, Henigsberg N et al (2010). Genome-wide association study of increasing suicidal ideation during antidepressant treatment in the GENDEP project. Pharmacogenomics J (in press).
Posner K, Oquendo MA, Gould M, Stanley B, Davies M (2007). Columbia Classification Algorithm of Suicide Assessment (C-CASA): classification of suicidal events in the FDA's pediatric suicidal risk analysis of antidepressants. Am J Psychiatry 164: 1035–1043.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909.
Rihmer Z, Akiskal H (2006). Do antidepressants t(h)reat(en) depressives? Toward a clinically judicious formulation of the antidepressant-suicidality FDA advisory in light of declining national suicide statistics from many countries. J Affect Disord 94: 3–13.
Rucci P, Frank E, Scocco P, Calugi S, Miniati M, Fagiolini A et al (2011). Treatment-emergent suicidal ideation during 4 months of acute management of unipolar major depression with SSRI pharmacotherapy or interpersonal psychotherapy in a randomized clinical trial. Depress Anxiety 28: 303–309.
Rush AJ, Bernstein IH, Trivedi MH, Carmody TJ, Wisniewski S, Mundt JC et al (2006). An evaluation of the quick inventory of depressive symptomatology and the Hamilton Rating Scale for depression: a sequenced treatment alternatives to relieve depression trial report. Biol Psychiatry 59: 493–501.
Rush AJ, Trivedi MH, Carmody TJ, Ibrahim HM, Markowitz JC, Keitner GI et al (2005). Self-reported depressive symptom measures: sensitivity to detecting change in a randomized, controlled trial of chronically depressed, nonpsychotic outpatients. Neuropsychopharmacology 30: 405–416.
Rush AJ, Trivedi MH, Ibrahim HM, Carmody TJ, Arnow B, Klein DN et al (2003). The 16-Item Quick Inventory of Depressive Symptomatology (QIDS), clinician rating (QIDS-C), and self-report (QIDS-SR): a psychometric evaluation in patients with chronic major depression. Biol Psychiatry 54: 573–583.
Seemuller F, Riedel M, Obermeier M, Bauer M, Adli M, Mundt C et al (2009). The controversial link between antidepressants and suicidality risks in adults: data from a naturalistic study on a large sample of in-patients with a major depressive episode. Int J Neuropsychopharmacol 12: 181–189.
Statham DJ, Heath AC, Madden PA, Bucholz KK, Bierut L, Dinwiddie SH et al (1998). Suicidal behaviour: an epidemiological and genetic study. Psychol Med 28: 839–855.
Stopkova P, Saito T, Papolos DF, Vevera J, Paclt I, Zukov I et al (2004). Identification of PIK3C3 promoter variant associated with bipolar disorder and schizophrenia. Biol Psychiatry 55: 981–988.
Szanto K, Mulsant BH, Houck PR, Dew MA, Dombrovski A, Pollock BG et al (2007). Emergence, persistence, and resolution of suicidal ideation during treatment of depression in old age. J Affect Disord 98: 153–161.
Tang K, Fu DJ, Julien D, Braun A, Cantor CR, Koster H (1999). Chip-based genotyping by mass spectrometry. Proc Natl Acad Sci USA 96: 10016–10020.
Tang R, Zhao X, Fang C, Tang W, Huang K, Wang L et al (2008). Investigation of variants in the promoter region of PIK3C3 in schizophrenia. Neurosci Lett 437: 42–44.
Teicher MH, Glod C, Cole JO (1990). Emergence of intense suicidal preoccupation during fluoxetine treatment. Am J Psychiatry 147: 207–210.
Trivedi MH, Rush AJ, Ibrahim HM, Carmody TJ, Biggs MM, Suppes T et al (2004). The Inventory of Depressive Symptomatology, Clinician Rating (IDS-C) and Self-Report (IDS-SR), and the Quick Inventory of Depressive Symptomatology, Clinician Rating (QIDS-C) and Self-Report (QIDS-SR) in public sector patients with mood disorders: a psychometric evaluation. Psychol Med 34: 73–82.
Valuck RJ, Libby AM, Orton HD, Morrato EH, Allen R, Baldessarini RJ (2007). Spillover effects on treatment of adult depression in primary care after FDA advisory on risk of pediatric suicidality with SSRIs. Am J Psychiatry 164: 1198–1205.
Wang L, Budolfson K, Wang F (2011). Pik3c3 deletion in pyramidal neurons results in loss of synapses, extensive gliosis and progressive neurodegeneration. Neuroscience 172: 427–442.
Westfall PH (1993). Resampling-based Multiple Testing. Wiley: New York.
Wigginton JE, Cutler DJ, Abecasis GR (2005). A note on exact tests of Hardy–Weinberg equilibrium. Am J Hum Genet 76: 887–893.
Wirshing WC, Van Putten T, Rosenberg J, Marder S, Ames D, Hicks-Gray T (1992). Fluoxetine, akathisia, and suicidality: is there a causal connection? Arch Gen Psychiatry 49: 580–581.
Zisook S, Trivedi MH, Warden D, Lebowitz B, Thase ME, Stewart JW et al (2009). Clinical correlates of the worsening or emergence of suicidal ideation during SSRI treatment of depression: An examination of citalopram in the STARD study. J Affect Disord 117: 63–73.
Acknowledgements
We thank M Rex-Haffner, S Damast, S Sauer, G Ernst-Jansen, E Kappelmann, M Hartung, and B Siegel for their excellent technical assistance. The study is supported by a grant of the Exzellenz-Stiftung of the Max Planck Society. This work has also been funded by the Federal Ministry of Education and Research (BMBF) in the framework of the National Genome Research Network (NGFN), FKZ 01GS0481.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Disclosures: Czamara, Klengel, Hennings, Lucae, Arolt: nothing to disclose.
Patent applications:
Menke, Binder, Holsboer inventors:
Means and methods for diagnosing predisposition for treatment-emergent suicidal ideation (TESI). European application number: 08016477.5; International application number: PCT/EP2009/061575.
Binder, Müller-Myhsok, Holsboer inventors:
FKBP5: a novel target for antidepressant therapy. International publication number: WO 2005/054500.
Polymorphisms in ABCB1 associated with a lack of clinical response to medicaments. International application number: PCT/EP2005/005194.
Other disclosures:
Elisabeth B Binder: Currently grant support NIMH, Doris Duke Charitable foundation, Behrens-Weise-Stiftung, and PharmaNeuroBoost.
Florian Holsboer: Founder and share holder: Affectis Pharmaceuticals and HolsboerMaschmeyer NeuroChemie GmbH.
Katharina Domschke: Katharina Domschke has received speaker fees from Pfizer, Lilly and Bristol-Myers Squibb, she is a consultant for Johnson & Johnson and has received funding by Astra Zeneca.
Bernhard Baune: Currently grant support from NHMRC and Astra Zeneca.
Additional information
Parts of the data have been presented at the Society of Biological Psychiatry, May 2009, Vancouver and at the Symposium of the AGNP, October 2009, Munich
Supplementary Information accompanies the paper on the Neuropsychopharmacology website
Supplementary information
PowerPoint slides
Rights and permissions
About this article
Cite this article
Menke, A., Domschke, K., Czamara, D. et al. Genome-Wide Association Study of Antidepressant Treatment-Emergent Suicidal Ideation. Neuropsychopharmacol 37, 797–807 (2012). https://doi.org/10.1038/npp.2011.257
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/npp.2011.257
Keywords
This article is cited by
-
Genome-wide significant regions in 43 Utah high-risk families implicate multiple genes involved in risk for completed suicide
Molecular Psychiatry (2020)
-
Genetic variation in the Estonian population: pharmacogenomics study of adverse drug effects using electronic health records
European Journal of Human Genetics (2019)
-
Gene expression associated with suicide attempts in US veterans
Translational Psychiatry (2017)
-
Towards understanding and predicting suicidality in women: biomarkers and clinical risk assessment
Molecular Psychiatry (2016)
-
Pharmacogenetics and Imaging–Pharmacogenetics of Antidepressant Response: Towards Translational Strategies
CNS Drugs (2016)