Abstract
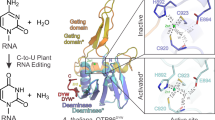
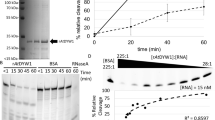
RNA editing is a post-transcriptional process that modifies the genetic information on RNA molecules. In flowering plants, RNA editing usually alters cytidine to uridine in plastids and mitochondria. The PLS-type pentatricopeptide repeat (PPR) protein and the multiple organellar RNA editing factor (MORF, also known as RNA editing factor interacting protein (RIP)) are two types of key trans-acting factors involved in this process. However, how they cooperate with one another remains unclear. Here, we have characterized the interactions between a designer PLS-type PPR protein (PLS)3PPR and MORF9, and found that RNA-binding activity of (PLS)3PPR is drastically increased on MORF9 binding. We also determined the crystal structures of (PLS)3PPR, MORF9 and the (PLS)3PPR–MORF9 complex. MORF9 binding induces significant compressed conformational changes of (PLS)3PPR, revealing the molecular mechanisms by which MORF9-bound (PLS)3PPR has increased RNA-binding activity. Similarly, increased RNA-binding activity is observed for the natural PLS-type PPR protein, LPA66, in the presence of MORF9. These findings significantly expand our understanding of MORF function in plant organellar RNA editing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Takenaka, M., Zehrmann, A., Verbitskiy, D., Hartel, B. & Brennicke, A. RNA editing in plants and its evolution. Annu. Rev. Genet. 47, 335–352 (2013).
Liu, Y. J., Xiu, Z. H., Meeley, R. & Tan, B. C. Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell 25, 868–883 (2013).
Huang, H. et al. RNA editing of the IQ domain in Cav1.3 channels modulates their Ca2+-dependent inactivation. Neuron 73, 304–316 (2012).
Chiu, Y. L. et al. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435, 108–114 (2005).
Zhu, Q. et al. The Arabidopsis thaliana RNA editing factor SLO2, which affects the mitochondrial electron transport chain, participates in multiple stress and hormone responses. Mol. Plant 7, 290–310 (2014).
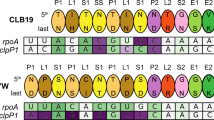
Chateigner-Boutin, A. L. et al. CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 56, 590–602 (2008).
Sosso, D. et al. PPR2263, a DYW-Subgroup Pentatricopeptide repeat protein, is required for mitochondrial nad5 and cob transcript editing, mitochondrion biogenesis, and maize growth. Plant Cell 24, 676–691 (2012).
Hwang, T. et al. Dynamic regulation of RNA editing in human brain development and disease. Nat. Neurosci. 19, 1093–1099 (2016).
Chen, L. et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 19, 209–216 (2013).
Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349 (2010).
Mehta, A. & Driscoll, D. M. Identification of domains in apobec-1 complementation factor required for RNA binding and apolipoprotein-B mRNA editing. RNA 8, 69–82 (2002).
Covello, P. S. & Gray, M. W. RNA editing in plant mitochondria. Nature 341, 662–666 (1989).
Gualberto, J. M., Lamattina, L., Bonnard, G., Weil, J. H. & Grienenberger, J. M. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature 341, 660–662 (1989).
Hiesel, R., Wissinger, B., Schuster, W. & Brennicke, A. RNA editing in plant mitochondria. Science 246, 1632–1634 (1989).
Hoch, B., Maier, R. M., Appel, K., Igloi, G. L. & Kossel, H. Editing of a chloroplast mRNA by creation of an initiation codon. Nature 353, 178–180 (1991).
Barkan, A. & Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442 (2014).
Bentolila, S., Oh, J., Hanson, M. R. & Bukowski, R. Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 9, e1003584 (2013).
Pyo, Y. J., Kwon, K. C., Kim, A. & Cho, M. H. Seedling Lethal1, a pentatricopeptide repeat protein lacking an E/E+ or DYW domain in Arabidopsis, is involved in plastid gene expression and early chloroplast development. Plant Physiol. 163, 1844–1858 (2013).
Lurin, C. et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103 (2004).
Yin, P. et al. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504, 168–171 (2013).
Shen, C. et al. Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 7, 11285 (2016).
Ke, J. et al. Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat. Struct. Mol. Biol. 20, 1377–1382 (2013).
Coquille, S. et al. An artificial PPR scaffold for programmable RNA recognition. Nat. Commun. 5, 5729 (2014).
Gully, B. S. et al. The design and structural characterization of a synthetic pentatricopeptide repeat protein. Acta Crystallogr. D 71, 196–208 (2015).
Takenaka, M. et al. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl Acad. Sci. USA 109, 5104–5109 (2012).
Bentolila, S. et al. RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc. Natl Acad. Sci. USA 109, E1453–E1461 (2012).
Cheng, S. et al. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 85, 532–547 (2016).
Barkan, A. et al. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8, e1002910 (2012).
Yagi, Y., Hayashi, S., Kobayashi, K., Hirayama, T. & Nakamura, T. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS ONE 8, e57286 (2013).
Holm, L., Kaariainen, S., Rosenstrom, P. & Schenkel, A. Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781 (2008).
Glass, F., Hartel, B., Zehrmann, A., Verbitskiy, D. & Takenaka, M. MEF13 requires MORF3 and MORF8 for RNA editing at eight targets in mitochondrial mRNAs in Arabidopsis thaliana. Mol. Plant 8, 1466–1477 (2015).
Kotera, E., Tasaka, M. & Shikanai, T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433, 326–330 (2005).
Shen, C. et al. Specific RNA recognition by designer pentatricopeptide repeat protein. Mol. Plant 8, 667–670 (2015).
Cai, W. et al. LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis. Plant Physiol. 150, 1260–1271 (2009).
Spahr, H. et al. SLIRP stabilizes LRPPRC via an RRM-PPR protein interface. Nucleic Acids Res. 44, 6868–6882 (2016).
Sun, T. et al. An RNA recognition motif-containing protein is required for plastid RNA editing in Arabidopsis and maize. Proc. Natl Acad. Sci. USA 110, E1169–E1178 (2013).
Shi, X., Hanson, M. R. & Bentolila, S. Two RNA recognition motif-containing proteins are plant mitochondrial editing factors. Nucleic Acids Res. 43, 3814–3825 (2015).
Zhang, F. et al. Tetrapyrrole biosynthetic enzyme protoporphyrinogen IX oxidase 1 is required for plastid RNA editing. Proc. Natl Acad. Sci. USA 111, 2023–2028 (2014).
Sun, T. et al. A zinc finger motif-containing protein is essential for chloroplast RNA editing. PLoS Genet. 11, e1005028 (2015).
Boussardon, C. et al. The cytidine deaminase signature HxE(x)nCxxC of DYW1 binds zinc and is necessary for RNA editing of ndhD-1. New Phytol. 203, 1090–1095 (2014).
Okuda, K. et al. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell 21, 146–156 (2009).
Wagoner, J. A., Sun, T., Lin, L. & Hanson, M. R. Cytidine deaminase motifs within the DYW domain of two pentatricopeptide repeat-containing proteins are required for site-specific chloroplast RNA editing. J. Biol. Chem. 290, 2957–2968 (2015).
Boussardon, C. et al. Two interacting proteins are necessary for the editing of the NdhD-1 site in Arabidopsis plastids. Plant Cell 24, 3684–3694 (2012).
Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. Weblogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Bendtsen, J. D., Nielsen, H., von Heijne, G. & Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 (2004).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002).
Acknowledgements
We thank the staff from BL17U/BL19U1/beamline of the National Centre for Protein Sciences Shanghai (NCPSS) at the Shanghai Synchrotron Radiation Facility for assistance during data collection, and research associates at the Centre for Protein Research, Huazhong Agricultural University, for technical support. This work was supported by funds from the Ministry of Science and Technology (grant 2015CB910900), the Fok Ying-Tong Education Foundation (grant 151021), the National Science Foundation of China (grant 91535301), the Fundamental Research Funds for the Central Universities (programme no. 2014PY026, no. 2015PY219 and no. 2014JQ001), Huazhong Agricultural University Scientific & Technological Self-innovation Foundation (programme no. 2013RC013) and the China Postdoctoral Science Foundation (grant 2015M572163).
Author information
Authors and Affiliations
Contributions
J.Y., Q.Z., R.L., T.Z. and P.Y. designed all experiments. J.Y., Q.Z., L.L. and F.R. performed protein expression, purification and crystallization. Z.G. determined all of the structures. J.Y., Q.Z. and Q.W. carried out biochemical assays. J.Y. and P.Y. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1–10, Supplementary Table 1. (PDF 1690 kb)
Rights and permissions
About this article
Cite this article
Yan, J., Zhang, Q., Guan, Z. et al. MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing. Nature Plants 3, 17037 (2017). https://doi.org/10.1038/nplants.2017.37
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2017.37
This article is cited by
-
Gene expression and physiological roles of post-transcriptional editing in plant organellar systems
Theoretical and Experimental Plant Physiology (2024)
-
The dicot homolog of maize PPR103 carries a C-terminal DYW domain and may have a role in C-to-U editing of some chloroplast RNA transcripts
Plant Molecular Biology (2024)
-
A ribonuclease activity linked to DYW1 in vitro is inhibited by RIP/MORF proteins
Scientific Reports (2023)
-
U-to-C RNA editing by synthetic PPR-DYW proteins in bacteria and human culture cells
Communications Biology (2022)
-
DYW domain structures imply an unusual regulation principle in plant organellar RNA editing catalysis
Nature Catalysis (2021)