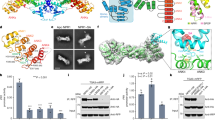
Abstract
The plant-specific GAI, RGA and SCR (GRAS) family proteins play critical roles in plant development and signalling. Two GRAS proteins, SHORT-ROOT (SHR) and SCARECROW (SCR), cooperatively direct asymmetric cell division and the patterning of root cell types by transcriptional control in conjunction with BIRD/INDETERMINATE DOMAIN (IDD) transcription factors, although precise details of these specific interactions and actions remain unknown. Here, we present the crystal structures of the SHR–SCR binary and JACKDAW (JKD)/IDD10–SHR–SCR ternary complexes. Each GRAS domain comprises one α/β core subdomain with an α-helical cap that mediates heterodimerization by forming an intermolecular helix bundle. The α/β core subdomain of SHR forms the BIRD binding groove, which specifically recognizes the zinc fingers of JKD. We identified a conserved SHR-binding motif in 13 BIRD/IDD transcription factors. Our results establish a structural basis for GRAS–GRAS and GRAS–BIRD interactions and provide valuable clues towards our understanding of these regulators, which are involved in plant-specific signalling networks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Pauluzzi, G. et al. Surfing along the root ground tissue gene network. Dev. Biol. 365, 14–22 (2012).
Petricka, J. J., Winter, C. M. & Benfey, P. N. Control of Arabidopsis root development. Annu. Rev. Plant Biol. 63, 563–590 (2012).
Benfey, P. N. et al. Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119, 57–70 (1993).
Scheres, B. et al. Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121, 53–62 (1995).
Di Laurenzio, L. et al. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423–433 (1996).
Helariutta, Y. et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567 (2000).
Sabatini, S., Heidstra, R., Wildwater, M. & Scheres, B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17, 354–358 (2003).
Cui, H., Kong, D., Liu, X. & Hao, Y. SCARECROW, SCR-LIKE 23 and SHORT-ROOT control bundle sheath cell fate and function in Arabidopsis thaliana. Plant J. 78, 319–327 (2014).
Nakajima, K., Sena, G., Nawy, T. & Benfey, P. N. Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413, 307–311 (2001).
Heidstra, R., Welch, D. & Scheres, B. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 18, 1964–1969 (2004).
Cui, H. et al. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316, 421–425 (2007).
Levesque, M. P. et al. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 4, e143 (2006).
Colasanti, J. et al. The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7, 158 (2006).
Sozzani, R. et al. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466, 128–132 (2010).
Welch, D. et al. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 21, 2196–2204 (2007).
Long, Y. et al. Arabidopsis BIRD zinc finger proteins jointly stabilize tissue boundaries by confining the cell fate regulator SHORT-ROOT and contributing to fate specification. Plant Cell 27, 1185–1199 (2015).
Moreno-Risueno, M. A. et al. Transcriptional control of tissue formation throughout root development. Science 350, 426–430 (2015).
Pysh, L. D., Wysocka-Diller, J. W., Camilleri, C., Bouchez, D. & Benfey, P. N. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18, 111–119 (1999).
Bolle, C. The role of GRAS proteins in plant signal transduction and development. Planta 218, 683–692 (2004).
Murase, K., Hirano, Y., Sun, T. P. & Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463 (2008).
Sun, X., Jones, W. T. & Rikkerink, E. H. GRAS proteins: the versatile roles of intrinsically disordered proteins in plant signalling. Biochem. J. 442, 1–12 (2012).
Gallagher, K. L. & Benfey, P. N. Both the conserved GRAS domain and nuclear localization are required for SHORT-ROOT movement. Plant J. 57, 785–797 (2009).
Martin, J. L. & McMillan, F. M. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 12, 783–793 (2002).
Joshi, C. P. & Chiang, V. L. Conserved sequence motifs in plant S-adenosyl-L-methionine-dependent methyltransferases. Plant Mol. Biol. 37, 663–674 (1998).
Pabo, C. O., Peisach, E. & Grant, R. A. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 70, 313–340 (2001).
Kozaki, A., Hake, S. & Colasanti, J. The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res. 32, 1710–1720 (2004).
Ogasawara, H., Kaimi, R., Colasanti, J. & Kozaki, A. Activity of transcription factor JACKDAW is essential for SHR/SCR-dependent activation of SCARECROW and MAGPIE and is modulated by reciprocal interactions with MAGPIE, SCARECROW and SHORT ROOT. Plant Mol. Biol. 77, 489–499 (2011).
Pavletich, N. P. & Pabo, C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1Å. Science 252, 809–817 (1991).
Hirsch, S. et al. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21, 545–557 (2009).
Li, S. et al. Crystal structure of the GRAS domain of SCARECROW-LIKE 7 in Oryza sativa. Plant Cell 28, 1025–1034 (2016).
Hamiaux, C. et al. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22, 2032–2036 (2012).
Kagiyama, M. et al. Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 18, 147–160 (2013).
Yoshida, H. et al. DELLA protein functions as a transcriptional activator through the DNA binding of the INDETERMINATE DOMAIN family proteins. Proc. Natl Acad. Sci. USA 111, 7861–7866 (2014).
Fukazawa, J. et al. DELLAs function as coactivators of GAI-ASSOCIATED FACTOR1 in regulation of gibberellin homeostasis and signaling in Arabidopsis. Plant Cell 26, 2920–2938 (2014).
Doublié, S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 (1997).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Schneider, T. R. & Sheldrick, G. M. Substructure solution with SHELXD. Acta Crystallogr. D 58, 1772–1779 (2002).
Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Holm, L. & Park, J. Dalilite workbench for protein structure comparison. Bioinformatics 16, 566–567 (2000).
Nagai, T. et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 (2002).
Shimura, H. et al. A strategy for screening an inhibitor of viral silencing suppressors, which attenuates symptom development of plant viruses. FEBS Lett. 582, 4047–4052 (2008).
Acknowledgements
We thank R. Kurata for technical support in performing the MALDI-TOF MS analysis. We appreciate access to BL41XU and BL44XU beamlines at the SPring-8 synchrotron facility for the provision of synchrotron data collection facilities (proposal nos. 2013A6844, 2013B1288, 2013B6844, 2014A 1283 and 2014A6944). The cDNAs were provided by RIKEN BRC through the National Bio-Resource Project of the MEXT, Japan. This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas ‘Structural Cell Biology’ (to T.H.), a Grant-in-Aid for Scientific Research (C) (to Y.H.) from MEXT, Japan, the Core Research for Evolutionary Science and Technology (CREST), the Japan Science and Technology Agency (to Y.H.) and the National Institutes of Health, United States (R01 GM100051 to T.-p.S.).
Author information
Authors and Affiliations
Contributions
Y.H. and T.H. conceived and designed the project. Y.H. was responsible for construct design for the protein preparations. M.S., Y.H. and M.N. prepared the DNA constructs. Y.H., M.N. and T.S. performed protein biochemistry, crystallization and X-ray data collection. Y.H. solved and refined the structures. K.M. performed in vivo binding assay supervised by S.T., Y.H., M.N., T.S. and T.H. interpreted data. Y.H., T.-p.S. and T.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information 1–3, Supplementary Table 1, Supplementary Figures 1–12, Supplementary Figure Legends 1–12. (PDF 4184 kb)
Rights and permissions
About this article
Cite this article
Hirano, Y., Nakagawa, M., Suyama, T. et al. Structure of the SHR–SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD. Nature Plants 3, 17010 (2017). https://doi.org/10.1038/nplants.2017.10
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2017.10
This article is cited by
-
Functional characterization of plant specific Indeterminate Domain (IDD) transcription factors in tomato (Solanum lycopersicum L.)
Scientific Reports (2024)
-
The master growth regulator DELLA binding to histone H2A is essential for DELLA-mediated global transcription regulation
Nature Plants (2023)
-
SHORT-ROOT stabilizes PHOSPHATE1 to regulate phosphate allocation in Arabidopsis
Nature Plants (2022)
-
Genome-wide investigation of the GRAS transcription factor family in foxtail millet (Setaria italica L.)
BMC Plant Biology (2021)
-
SCARECROW-LIKE3 regulates the transcription of gibberellin-related genes by acting as a transcriptional co-repressor of GAI-ASSOCIATED FACTOR1
Plant Molecular Biology (2021)