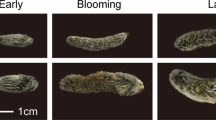
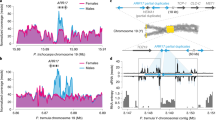
Abstract
Separate sexes and sex-biased gene expression have repeatedly evolved in animals and plants, but the underlying changes in gene expression remain unknown. Here, we studied a pair of plant species, one in which separate sexes and sex chromosomes evolved recently and one which maintained hermaphrodite flowers resembling the ancestral state, to reconstruct expression changes associated with the evolution of dioecy. We found that sex-biased gene expression has evolved in autosomal and sex-linked genes in the dioecious species. Most expression changes relative to hermaphrodite flowers occurred in females rather than males, with higher and lower expression in females leading to female-biased and male-biased expression, respectively. Expression changes were more common in genes located on the sex chromosomes than the autosomes and led to feminization of the X chromosome and masculinization of the Y chromosome. Our results support a scenario in which sex-biased gene expression evolved during the evolution of dioecy to resolve intralocus sexual conflicts over the allocation of resources.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ellegren, H. & Parsch, J. The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 8, 689–698 (2007).
Mank, J. E. Sex chromosomes and the evolution of sexual dimorphism: lessons from the genome. Am. Nat. 173, 141–150 (2009).
Stewart, A. D., Pischedda, A. & Rice, W. R. Resolving intralocus sexual conflict: genetic mechanisms and time frame. J. Hered. 101(Suppl. 1), S94–S99 (2010).
van Doorn, G. S. Intralocus sexual conflict. Ann. NY Acad. Sci. 1168, 52–71 (2009).
Bonduriansky, R. & Chenoweth, S. F. Intralocus sexual conflict. Trends Ecol. Evol. 24, 280–288 (2009).
Reinius, B. et al. Abundance of female-biased and paucity of male-biased somatically expressed genes on the mouse X-chromosome. BMC Genomics 13, 607 (2012).
Meisel, R. P., Malone, J. H. & Clark, A. G. Disentangling the relationship between sex-biased gene expression and X-linkage. Genome Res. 22, 1255–1265 (2012).
Ellegren, H. Emergence of male-biased genes on the chicken Z-chromosome: sex-chromosome contrasts between male and female heterogametic systems. Genome Res. 21, 2082–2086 (2011).
Kang, H. J. et al. Spatio-temporal transcriptome of the human brain. Nature 478, 483–489 (2011).
Lipinska, A. et al. Sexual dimorphism and the evolution of sex-biased gene expression in the brown alga Ectocarpus. Mol. Biol. Evol. 32, 1581–1597 (2015).
Harkess, A. et al. Sex-biased gene expression in dioecious garden asparagus (Asparagus officinalis). New Phytol. 207, 883–892 (2015).
Robinson, K. M. et al. Populus tremula (European aspen) shows no evidence of sexual dimorphism. BMC Plant Biol. 14, 276 (2014).
Zemp, N., Tavares, R. & Widmer, A. Fungal infection induces sex-specific transcriptional changes and alters sexual dimorphism in the dioecious plant Silene latifolia. PLoS Genet. 11, e1005536 (2015).
Barrett, S. C. H. & Hough, J. Sexual dimorphism in flowering plants. J. Exp. Bot. 64, 67–82 (2013).
Bernasconi, G. et al. Silene as a model system in ecology and evolution. Heredity 103, 5–14 (2009).
Bergero, R., Qiu, S. & Charlesworth, D. Gene loss from a plant sex chromosome system. Curr. Biol. 25, 1234–1240 (2015).
Chibalina, M. V. & Filatov, D. A. Plant Y chromosome degeneration is retarded by haploid purifying selection. Curr. Biol. 21, 1475–1479 (2011).
Marais, G. A. B. et al. Evidence for degeneration of the Y chromosome in the dioecious plant Silene latifolia. Curr. Biol. 18, 545–549 (2008).
Nicolas, M. et al. A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biol. 3, 47–56 (2005).
Papadopulos, A. S. T., Chester, M., Ridout, K. & Filatov, D. A. Rapid Y degeneration and dosage compensation in plant sex chromosomes. Proc. Natl Acad. Sci. USA 112, 13021–13026 (2015).
Mrackova, M. et al. Independent origin of sex chromosomes in two species of the genus Silene. Genetics 179, 1129–1133 (2008).
Delph, L. F., Knapczyk, F. N. & Taylor, D. R. Among-population variation and correlations in sexually dimorphic traits of Silene latifolia. J. Evol. Biol. 15, 1011–1020 (2002).
Delph, L. F., Arntz, A. M., Scotti-Saintagne, C. & Scotti, I. The genomic architecture of sexual dimorphism in the dioecious plant Silene latifolia. Evolution 64, 2873–2886 (2010).
Marais, G. A. B. et al. Multiple nuclear gene phylogenetic analysis of the evolution of dioecy and sex chromosomes in the genus Silene. PLoS ONE 6, e21915 (2011).
Rautenberg, A., Sloan, D. B., Alden, V. & Oxelman, B. Phylogenetic relationships of Silene multinervia and Silene section Conoimorpha (Caryophyllaceae). Syst. Bot. 37, 226–237 (2012).
Bergero, R. & Charlesworth, D. Preservation of the Y transcriptome in a 10-million-year-old plant sex chromosome system. Curr. Biol. 21, 1470–1474 (2011).
Muyle, A. et al. SEX-DETector: a probabilistic approach to study sex chromosomes in non-model organisms. Genome Biol. Evol. 8, 2530–2543 (2016).
Mank, J. E. Sex chromosome dosage compensation: definitely not for everyone. Trends Genet. 29, 677–683 (2013).
Muyle, A. et al. Rapid de novo evolution of X chromosome dosage compensation in Silene latifolia, a plant with young sex chromosomes. PLoS Biol. 10, e1001308 (2012).
Hough, J., Hollister, J. D., Wang, W., Barrett, S. C. H. & Wright, S. I. Genetic degeneration of old and young Y chromosomes in the flowering plant Rumex hastatulus. Proc. Natl Acad. Sci. USA 111, 7713–7718 (2014).
Moore, J. C. & Pannell, J. R. Sexual selection in plants. Curr. Biol. 21, R176–R182 (2011).
Delph, L. F. & Herlihy, C. R. Sexual, fecundity, and viability selection on flower size and number in a sexually dimorphic plant. Evolution 66, 1154–1166 (2012).
Vicoso, B., Kaiser, V. B. & Bachtrog, D. Sex-biased gene expression at homomorphic sex chromosomes in emus and its implication for sex chromosome evolution. Proc. Natl Acad. Sci. USA 110, 6453–6458 (2013).
Jordan, C. Y. & Connallon, T. Sexually antagonistic polymorphism in simultaneous hermaphrodites. Evolution 68, 3555–3569 (2014).
Moghadam, H. K., Pointer, M. A., Wright, A. E., Berlin, S. & Mank, J. E. W chromosome expression responds to female-specific selection. Proc. Natl Acad. Sci. USA 109, 8207–8211 (2012).
Ometto, L., Shoemaker, D., Ross, K. G. & Keller, L. Evolution of gene expression in fire ants: the effects of developmental stage, caste, and species. Mol. Biol. Evol. 28, 1381–1392 (2011).
Connalon, T. & Knowles, L. L. Intergenomic conflict revealed by patterns of sex-biased gene expression. Trends Genet. 21, 495–499 (2005).
Disteche, C. M. Dosage compensation of the sex chromosomes. Annu. Rev. Genet. 46, 537–560 (2012).
Zhou, Q. & Bachtrog, D. Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science 337, 341–345 (2012).
Skaletsky, H. et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 (2003).
Bellott, D. W. et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508, 494–499 (2014).
Cortez, D. et al. Origins and functional evolution of Y chromosomes across mammals. Nature 508, 488–493 (2014).
Charlesworth, B. & Charlesworth, D. The degeneration of Y chromosomes. Phil. Trans. R. Soc. Lond. B Biol. Sci. 355, 1563–1572 (2000).
Bachtrog, D. Y-chromosome evolution emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 14, 113–124 (2013).
Asikainen, E. & Mutikainen, P. Pollen and resource limitation in a gynodioecious species. Am. J. Bot. 92, 487–494 (2005).
Darwin, C. The Different Forms of Flowers on Plants of the Same Species. (John Murray, 1877).
Innocenti, P. & Morrow, E. H. The sexually antagonistic genes of Drosophila melanogaster. PLoS Biol. 8, e1000335 (2010).
Zhou, Q. & Bachtrog, D. Chromosome-wide gene silencing initiates Y degeneration in Drosophila. Curr. Biol. 22, 522–525 (2012).
Matsunaga, S. et al. Isolation and developmental expression of male reproductive organ-specific genes in a dioecious campion, Melandrium album (Silene latifolia). Plant J. 10, 679–690 (1996).
Charlesworth, B. & Charlesworth, D. A model for the evolution of dioecy and gynodioecy. Am. Nat. 112, 975–997 (1978).
Acknowledgements
This work was funded through SNF projects 141260 and 160123 to A.W. and the French National Research Agency ANR-11-BSV7-013-03 to G.A.B.M. We thank C. Michel for laboratory support, S. Zoller from the ETH Zurich Genetic Diversity Centre (GDC) for assembling the reference transcriptome and members of the Plant Ecological Genetics group for insightful discussions. We thank M.C. Fischer for providing pictures of S. latifolia and M. Frei for taking care of all plants. Data produced and analysed in this paper were generated in collaboration with the GDC.
Author information
Authors and Affiliations
Contributions
Designed the experiments and formulated predictions: N.Z., A.W., G.A.B.M., D.C.; performed the experiments: N.Z.; analysed the data: N.Z., R.T., A.M.; wrote the manuscript: N.Z., A.W., D.C., G.A.B.M., R.T., A.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Methods, Supplementary References, Supplementary Figures 1–9, Supplementary Tables 1–3 (PDF 1795 kb)
Rights and permissions
About this article
Cite this article
Zemp, N., Tavares, R., Muyle, A. et al. Evolution of sex-biased gene expression in a dioecious plant. Nature Plants 2, 16168 (2016). https://doi.org/10.1038/nplants.2016.168
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2016.168
This article is cited by
-
Gender specific SNP markers in Coscinium fenestratum (Gaertn.) Colebr. for resource augmentation
Molecular Biology Reports (2024)
-
Physiological aspects of sex differences and Haldane’s rule in Rumex hastatulus
Scientific Reports (2022)
-
Selection drives convergent gene expression changes during transitions to co-sexuality in haploid sexual systems
Nature Ecology & Evolution (2022)
-
Sex-biased genes and metabolites explain morphologically sexual dimorphism and reproductive costs in Salix paraplesia catkins
Horticulture Research (2021)
-
Genomic imprinting mediates dosage compensation in a young plant XY system
Nature Plants (2018)