Abstract
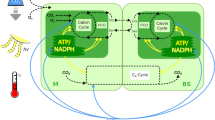
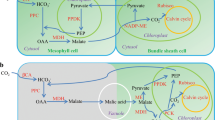
C4 photosynthesis acts as a carbon concentrating mechanism that leads to large increases in photosynthetic efficiency. The C4 pathway is found in more than 60 plant lineages1 but the molecular enablers of this evolution are poorly understood. In particular, it is unclear how non-photosynthetic proteins in the ancestral C3 system have repeatedly become strongly expressed and integrated into photosynthesis gene regulatory networks in C4 leaves. Here, we provide clear evidence that in C3 leaves, genes encoding key enzymes of the C4 pathway are already co-regulated with photosynthesis genes and are controlled by both light and chloroplast-to-nucleus signalling. In C4 leaves this regulation becomes increasingly dependent on the chloroplast. We propose that regulation of C4 cycle genes by light and the chloroplast in the ancestral C3 state has facilitated the repeated evolution of the complex and convergent C4 trait.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Sage, R. F., Christin, P.-A. & Edwards, E. J. The C4 plant lineages of planet Earth. J. Exp. Bot. 62, 3171–3181 (2011).
Jiao, Y., Lau, O. S. & Deng, X. W. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8, 217–230 (2007).
Chan, K. X., Phua, S. Y., Crisp, P., McQuinn, R. & Pogson, B. J. Learning the languages of the chloroplast: retrograde signaling and beyond. Annu. Rev. Plant Biol. 67, 25–53 (2016).
Aubry, S., Kelly, S., Kümpers, B. M. C., Smith-Unna, R. D. & Hibberd, J. M. Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Genet. 10, e1004365 (2014).
Kulahoglu, C. et al. Comparative transcriptome atlases reveal altered gene expression modules between two Cleomaceae C3 and C4 plant species. Plant Cell 26, 3243–3260 (2014).
Christin, P.-A. et al. Parallel recruitment of multiple genes into C4 photosynthesis. Genome Biol. Evol. 5, 2174–2187 (2013).
Hibberd, J. M. & Covshoff, S. The regulation of gene expression required for C4 photosynthesis. Annu. Rev. Plant Biol. 61, 181–207 (2010).
Eastmond, P. J. et al. Arabidopsis uses two gluconeogenic gateways for organic acids to fuel seedling establishment. Nat. Commun. 6, 6659 (2015).
Marshall, D. M. et al. Cleome, a genus closely related to Arabidopsis, contains species spanning a developmental progression from C3 to C4 photosynthesis. Plant J. 51, 886–896 (2007).
Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
The gene ontology consortium. Gene ontology consortium: going forward. Nucleic Acids Res. 43, D1049–D1056 (2015).
Ma, L. et al. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13, 2589–2607 (2001).
Covshoff, S. et al. Deregulation of maize C4 photosynthetic development in a mesophyll cell-defective mutant. Plant Physiol. 146, 1469–1481 (2008).
Tamada, Y. et al. Effect of photooxidative destruction of chloroplasts on the expression of nuclear genes for C4 photosynthesis and for chloroplast biogenesis in maize. J. Plant Physiol. 160, 3–8 (2003).
Charron, J.-B. F., He, H., Elling, A. A. & Deng, X. W. Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21, 3732–3748 (2009).
Rockwell, N. C., Su, Y.-S. & Lagarias, J. C. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858 (2006).
Lee, J. et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749 (2007).
Leivar, P. & Quail, P. H. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28 (2011).
Chen, F. et al. Arabidopsis phytochrome A directly targets numerous promoters for individualized modulation of genes in a wide range of pathways. Plant Cell 26, 1949–1966 (2014).
Tepperman, J. M. et al. Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J. 38, 725–739 (2004).
Shikata, H. et al. Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc. Natl Acad. Sci. USA 111, 18781–18786 (2014).
Zhang, H. et al. Genome-wide mapping of the HY5-mediated genenetworks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65, 346–358 (2011).
Toledo-Ortiz, G. et al. The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 10, e1004416 (2014).
Oh, E., Zhu, J.-Y. & Wang, Z.-Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802–809 (2012).
Oh, E. et al. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21, 403–419 (2009).
Zhang, Y. et al. A quartet of PIF bHLH factors provides a transcriptionally centered signaling Hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9, e1003244 (2013).
Ruckle, M. E., DeMarco, S. M. & Larkin, R. M. Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19, 3944–3960 (2007).
Kajala, K. et al. Multiple Arabidopsis genes primed for recruitment into C4 photosynthesis. Plant J. 69, 47–56 (2012).
Brown, N. J. et al. Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science 331, 1436–1439 (2011).
John, C. R., Smith-Unna, R. D., Woodfield, H., Covshoff, S. & Hibberd, J. M. Evolutionary convergence of cell-specific gene expression in independent lineages of C4 grasses. Plant Physiol. 165, 62–75 (2014).
McCormac, A. C. & Terry, M. J. Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J. 32, 549–559 (2002).
Pfaffl, M. W., Horgan, G. W. & Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, e36–e36 (2002).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Patro, R., Duggal, G. & Kingsford, C. Accurate, fast, and model-aware transcript expression quantification with salmon. bioRxiv http://dx.doi.org/10.1101/021592 (2015).
Hardcastle, T. J. & Kelly, K. A. Bayseq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 11, 422 (2010).
Hardcastle, T. J. bayseq: empirical Bayesian analysis of patterns of differential expression in count data. R package version 2.4.1 http://bioconductor.org/packages/baySeq/ (2012).
Carlson, M. org.At.tair.db: genome wide annotation for Arabidopsishttp://bioconductor.org/packages/org.At.tair.db/ (2015).
Alexa, A. & Rahnenfuhrer, J. topGO: enrichment analysis for Gene Ontology http://bioconductor.org/packages/topGO/ (2010).
RStudio Team. RStudio: Integrated Development Environment for R http://www.rstudio.com/ (2015).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2009); http://ggplot2.org/book/
Chen, F. et al. Photoreceptor partner FHY1 has an independent role in gene modulation and plant development under far-red light. Proc. Natl Acad. Sci. USA 111, 11888–11893 (2014).
Ouyang, X. et al. Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23, 2514–2535 (2011).
Hornitschek, P. et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signalling. Plant J. 71, 699–711 (2012).
Zhiponova, M. et al. Helix-loop-helix/basic helix-loop-helix transcription factor network represses cell elongation in Arabidopsis through an apparent incoherent feed-forward loop. Proc. Natl Acad. Sci. USA 111, 2824–2829 (2014).
Acknowledgements
The work was funded by the European Union 3to4 project and Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/J011754/1. I.G.-M. was supported by the Amgen Foundation. Research on chloroplast signalling by M.J.T. was supported by BBSRC grant (BB/J018139/1).
Author information
Authors and Affiliations
Contributions
S.J.B., M.J.T. and J.M.H. designed the study. S.J.B., I.G.-M. and M.J.G.-G. carried out the experimental work. S.J.B. and C.B. conducted the bioinformatics. S.J.B., M.J.T. and J.M.H. interpreted results and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1–6, Supplementary Tables 1 and 2, Supplementary Legends 3–7, Supplementary References. (PDF 591 kb)
Supplementary Table 1
Supplementary Table 1 (XLSX 10 kb)
Supplementary Table 2
Supplementary Table 2 (XLSX 11 kb)
Supplementary Table 3
Supplementary Table 3 (XLSX 5579 kb)
Supplementary Table 4
Supplementary Table 4 (XLSX 7569 kb)
Supplementary Table 5
Supplementary Table 5 (XLSX 19 kb)
Supplementary Table 6
Supplementary Table 6 (XLSX 16 kb)
Supplementary Table 7
Supplementary Table 7 (XLSX 11 kb)
Rights and permissions
About this article
Cite this article
Burgess, S., Granero-Moya, I., Grangé-Guermente, M. et al. Ancestral light and chloroplast regulation form the foundations for C4 gene expression. Nature Plants 2, 16161 (2016). https://doi.org/10.1038/nplants.2016.161
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2016.161
This article is cited by
-
The genetics of convergent evolution: insights from plant photosynthesis
Nature Reviews Genetics (2019)
-
Wheat genomics: Seeds of C4 photosynthesis
Nature Plants (2016)