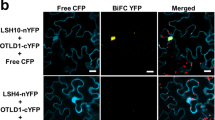
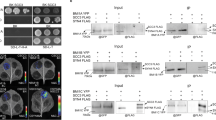
Abstract
Polycomb group (PcG) proteins form an epigenetic memory system in plants and animals, but interacting proteins are poorly known in plants. Here, we have identified Arabidopsis UBIQUITIN SPECIFIC PROTEASES (USP; UBP in plant and yeasts) 12 and 13 as partners of the plant-specific PcG protein LIKE HETEROCHROMATIN PROTEIN 1 (LHP1). UBP12 binds to chromatin of PcG target genes and is required for histone H3 lysine 27 trimethylation and repression of a subset of PcG target genes. Plants lacking UBP12 and UBP13 developed autonomous endosperm in the absence of fertilization. We have identified UBP12 and UBP13 as new proteins in the plant PcG regulatory network. UBP12 and UBP13 belong to an ancient gene family and represent plant homologues of metazoan USP7. We have found that Drosophila USP7 shares a function in heterochromatic gene repression with UBP12/13 and their homologue UBP26. In summary, we demonstrate that USP7-like proteins are essential for gene silencing in diverse genomic contexts.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Lewis, E. B. A gene complex controlling segmentation in Drosophila. Nature 276, 565–570 (1978).
Simon, J. A. & Kingston, R. E. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49, 808–824 (2013).
Derkacheva, M. & Hennig, L. Variations on a theme: Polycomb group proteins in plants. J. Exp. Bot. 65, 2769–2784 (2014).
Holec, S. & Berger, F. Polycomb group complexes mediate developmental transitions in plants. Plant Physiol. 158, 35–43 (2012).
Bratzel, F., Lopez-Torrejon, G., Koch, M., Del Pozo, J. C. & Calonje, M. Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 20, 1853–1859 (2010).
Yang, C. et al. VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol. 23, 1324–1329 (2013).
Blackledge, N. P. et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157, 1445–1459 (2014).
Calonje, M. PRC1 marks the difference in plant PcG repression. Mol. Plant 7, 459–471 (2014).
Cooper, S. et al. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 7, 1456–1470 (2014).
Kalb, R. et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 21, 569–571 (2014).
Sanchez-Pulido, L., Devos, D., Sung, Z. R. & Calonje, M. RAWUL: a new Ubiquitin-like domain in PRC1 Ring finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genomics 9, 308 (2008).
Xu, L. & Shen, W. H. Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr. Biol. 18, 1966–1971 (2008).
Turck, F. et al. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of Histone H3 Lysine 27. PLoS Genet. 3, 0855–0866 (2007).
Zhang, X. et al. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 14, 869–871 (2007).
Exner, V. et al. The chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 is essential for H3K27me3 binding and function during Arabidopsis development. PLoS ONE 4, e5335 (2009).
Kotake, T., Takada, S., Nakahigashi, K., Ohto, M. & Goto, K. Arabidopsis TERMINAL FLOWER 2 gene encodes a LIKE HETEROCHROMATIN PROTEIN 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol. 44, 555–564 (2003).
Libault, M. et al. The Arabidopsis LHP1 protein is a component of euchromatin. Planta 222, 910–925 (2005).
Derkacheva, M. et al. Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 32, 2073–2085 (2013).
Wang, Y., Gu, X., Yuan, W., Schmitz, R. J. & He, Y. Photoperiodic control of the floral transition through a distinct polycomb repressive complex. Dev. Cell 28, 727–736 (2014).
Makarova, K. S., Aravind, L. & Koonin, E. V. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem. Sci. 25, 50–52 (2000).
Yan, N., Doelling, J. H., Falbel, T. G., Durski, A. M. & Vierstra, R. D. The ubiquitin-specific protease family from Arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol 124, 1828–1843 (2000).
Liu, Y. et al. Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development. Plant J. 55, 844–856 (2008).
Ewan, R. et al. Deubiquitinating enzymes AtUBP12 and AtUBP13 and their tobacco homologue NtUBP12 are negative regulators of plant immunity. New Phytol. 191, 92–106 (2011).
Doelling, J. H. et al. The ubiquitin-specific protease subfamily UBP3/UBP4 is essential for pollen development and transmission in Arabidopsis. Plant Physiol. 145, 801–813 (2007).
Sridhar, V. V. et al. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447, 735–738 (2007).
Cui, X. et al. Ubiquitin-specific proteases UBP12 and UBP13 act in circadian clock and photoperiodic flowering regulation in Arabidopsis. Plant Physiol. 162, 897–906 (2013).
Shen, L. et al. The putative PRC1 RING-finger protein AtRING1A regulates flowering through repressing MADS AFFECTING FLOWERING genes in Arabidopsis. Development 141, 1303–1312 (2014).
Ohad, N. et al. A mutation that allows endosperm development without fertilization. Proc. Natl Acad. Sci. USA 93, 5319–5324 (1996).
Chaudhury, A. M. et al. Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 94, 4223–4228 (1997).
Köhler, C. et al. Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22, 4804–4814 (2003).
Guitton, A. E. et al. Identification of new members of FERTILISATION INDEPENDENT SEED Polycomb group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131, 2971–2981 (2004).
Pillot, M. et al. Embryo and endosperm inherit distinct chromatin and transcriptional states from the female gametes in Arabidopsis. Plant Cell 22, 307–320 (2010).
Wuest, S. E. et al. Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr. Biol. 20, 506–512 (2010).
Roszak, P. & Köhler, C. Polycomb group proteins are required to couple seed coat initiation to fertilization. Proc. Natl Acad. Sci USA 108, 20826–20831 (2011).
Mozgova, I. & Hennig, L. The polycomb group protein regulatory network. Annu. Rev. Plant Biol. 66, 269–296 (2015).
Scheuermann, J. C. et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465, 243–247 (2010).
Cotto-Rios, X. M., Bekes, M., Chapman, J., Ueberheide, B. & Huang, T. T. Deubiquitinases as a signaling target of oxidative stress. Cell Rep. 2, 1475–1484 (2012).
Lee, J. G., Baek, K., Soetandyo, N. & Ye, Y. Reversible inactivation of deubiquitinases by reactive oxygen species in vitro and in cells. Nat. Commun. 4, 1568 (2013).
Luo, M. et al. UBIQUITIN-SPECIFIC PROTEASE 26 is required for seed development and the repression of PHERES1 in Arabidopsis. Genetics 180, 229–236 (2008).
Schmitz, R. J., Tamada, Y., Doyle, M. R., Zhang, X. & Amasino, R. M. Histone H2B deubiquitination is required for transcriptional activation of FLOWERING LOCUS C and for proper control of flowering in Arabidopsis. Plant Physiol. 149, 1196–1204 (2009).
van der Knaap, J. A. et al. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol. Cell 17, 695–707 (2005).
Tartof, K. D., Hobbs, C. & Jones, M. A structural basis for variegating position effects. Cell 37, 869–878 (1984).
van der Knaap, J. A., Kozhevnikova, E., Langenberg, K., Moshkin, Y. M. & Verrijzer, C. P. Biosynthetic enzyme GMP synthetase cooperates with ubiquitin-specific protease 7 in transcriptional regulation of ecdysteroid target genes. Mol. Cell Biol. 30, 736–744 (2010).
Gaudin, V. et al. Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development 128, 4847–4858 (2001).
Cui, H. & Benfey, P. N. Interplay between SCARECROW, GA and LIKE HETEROCHROMATIN PROTEIN 1 in ground tissue patterning in the Arabidopsis root. Plant J. 58, 1016–1027 (2009).
Liu, C., Xi, W., Shen, L., Tan, C. & Yu, H. Regulation of floral patterning by flowering time genes. Dev. Cell 16, 711–722 (2009).
Latrasse, D. et al. Control of flowering and cell fate by LIF2, an RNA binding partner of the Polycomb complex component LHP1. PLoS One 6, e16592 (2011).
Takada, S. & Goto, K. TERMINAL FLOWER 2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN 1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15, 2856–2865 (2003).
Mylne, J. S. et al. LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc. Natl Acad. Sci. USA 103, 5012–5017 (2006).
Pengelly, A. R., Kalb, R., Finkl, K. & Muller, J. Transcriptional repression by PRC1 in the absence of H2A monoubiquitylation. Genes Dev. 29, 1487–1492 (2015).
Scheuermann, J. C., Gutierrez, L. & Muller, J. Histone H2A monoubiquitination and Polycomb repression: the missing pieces of the puzzle. Fly 6, 162–168 (2012).
Acknowledgements
We thank B. Liu (Uppsala) for help with protein expression, E. Savenkov (Uppsala) for providing plasmid pSITE–nYFP-N1-2b, M. Blatt (University of Glasgow) for providing a set of ubiquitin promoter-based vectors with fluorescent tags, X. Cui (Chinese Academy of Science, Beijing) for ubp12-2w ubp13-3 seeds and P. Verrijzer (Erasmus University Medical Centre, Rotterdam) for USP7 mutant flies. We thank TAIR and ABRC for mutant seeds and clones. We thank W. Gruissem for continuous support and providing access to infrastructure. This work was supported by grants from ETH Zurich, the Swedish Research Council and the Knut-and-Alice-Wallenberg Foundation.
Author information
Authors and Affiliations
Contributions
M.D., S.L., D.D.F., M.G., I.M., P.N. and M.T. performed the experiments; M.D., S.L., D.D.F., I.M. and P.N. analysed data; M.D., S.L., M.M., C.K. and L.H. planned the experiments; and M.D., M.M., C.K. and L.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Methods, Supplementary Figures, Supplementary Tables 1- 3, Supplementary References (PDF 1122 kb)
Supplementary Information
Supplementary Table 4 (XLSX 25 kb)
Rights and permissions
About this article
Cite this article
Derkacheva, M., Liu, S., Figueiredo, D. et al. H2A deubiquitinases UBP12/13 are part of the Arabidopsis polycomb group protein system. Nature Plants 2, 16126 (2016). https://doi.org/10.1038/nplants.2016.126
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2016.126
This article is cited by
-
The UBP5 histone H2A deubiquitinase counteracts PRCs-mediated repression to regulate Arabidopsis development
Nature Communications (2024)
-
The Function of Deubiquitinating Enzymes in Arabidopsis: Recent Progress of Ubiquitin-Specific Proteases (UBPs)
Journal of Plant Biology (2024)
-
Genome-Wide Identification, Characterization, and Expression Profiling of Eukaryotic-Specific UBP Family Genes in Brassica rapa
Journal of Plant Growth Regulation (2023)
-
Polycomb Repressive Complexes and Their Roles in Plant Developmental Programs, Particularly Floral Transition
Journal of Plant Biology (2023)
-
Plant deubiquitinases: from structure and activity to biological functions
Plant Cell Reports (2023)