Abstract
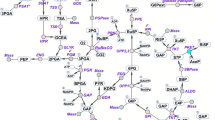
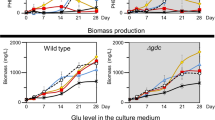
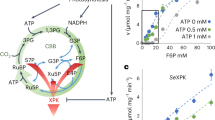
Central carbon metabolism in cyanobacteria comprises the Calvin–Benson–Bassham (CBB) cycle, glycolysis, the pentose phosphate (PP) pathway and the tricarboxylic acid (TCA) cycle. Redundancy in this complex metabolic network renders the rational engineering of cyanobacterial metabolism for the generation of biomass, biofuels and chemicals a challenge. Here we report the presence of a functional phosphoketolase pathway, which splits xylulose-5-phosphate (or fructose-6-phosphate) to acetate precursor acetyl phosphate, in an engineered strain of the model cyanobacterium Synechocystis (ΔglgC/xylAB), in which glycogen synthesis is blocked, and xylose catabolism enabled through the introduction of xylose isomerase and xylulokinase. We show that this mutant strain is able to metabolise xylose to acetate on nitrogen starvation. To see whether acetate production in the mutant is linked to the activity of phosphoketolase, we disrupted a putative phosphoketolase gene (slr0453) in the ΔglgC/xylAB strain, and monitored metabolic flux using 13C labelling; acetate and 2-oxoglutarate production was reduced in the light. A metabolic flux analysis, based on isotopic data, suggests that the phosphoketolase pathway metabolises over 30% of the carbon consumed by ΔglgC/xylAB during photomixotrophic growth on xylose and CO2. Disruption of the putative phosphoketolase gene in wild-type Synechocystis also led to a deficiency in acetate production in the dark, indicative of a contribution of the phosphoketolase pathway to heterotrophic metabolism. We suggest that the phosphoketolase pathway, previously uncharacterized in photosynthetic organisms, confers flexibility in energy and carbon metabolism in cyanobacteria, and could be exploited to increase the efficiency of cyanobacterial carbon metabolism and photosynthetic productivity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
McEwen, J. T., Machado, I. M. P., Connor, M. R. & Atsumi, S. Engineering Synechococcus elongatus PCC 7942 for continuous growth under diurnal conditions. Appl. Environ. Microbiol. 79, 1668–1675 (2013).
Lee, T. C. et al. Engineered xylose utilization enhances bio-products productivity in the cyanobacterium Synechocystis sp. PCC 6803. Metab. Eng. 30, 179–189 (2015).
Yan, C. L. & Xu, X. D. Bifunctional enzyme FBPase/SBPase is essential for photoautotrophic growth in cyanobacterium Synechocystis sp PCC 6803. Prog. Nat. Sci. 18, 149–153 (2008).
Bricker, T. M. et al. The malic enzyme is required for optimal photoautotrophic growth of Synechocystis sp. strain PCC 6803 under continuous light but not under a diurnal light regimen. J. Bacteriol. 186, 8144–8148 (2004).
Jansen, T. et al. Characterization of trophic changes and a functional oxidative pentose phosphate pathway in Synechocystis sp PCC 6803. Acta Physiol. Plant. 32, 511–518 (2010).
Xiong, W., Brune, D. & Vermaas, W. F. J. The gamma-aminobutyric acid shunt contributes to closing the tricarboxylic acid cycle in Synechocystis sp PCC 6803. Mol. Microbiol. 93, 786–796 (2014).
Gu, Z. L. et al. Role of duplicate genes in genetic robustness against null mutations. Nature 421, 63–66 (2003).
Valverde, F. et al. Simultaneous occurrence of two different glyceraldehyde-3-phosphate dehydrogenases in heterocystous N2-fixing cyanobacteria. Biochem. Biophys. Res. Commun. 283, 356–363 (2001).
Holtman, C. K. et al. High-throughput functional analysis of the Synechococcus elongatus PCC 7942 genome. DNA Res. 12, 103–115 (2005).
Tyo, K. E., Jin, Y. S., Espinoza, F. A. & Stephanopoulos, G. Identification of gene disruptions for increased poly-3-hydroxybutyrate accumulation in Synechocystis PCC 6803. Biotechnol. Prog. 25, 1236–1243 (2009).
Yang, C., Hua, Q. & Shimizu, K. Metabolic flux analysis in Synechocystis using isotope distribution from 13C-labeled glucose. Metab. Eng. 4, 202–216 (2002).
Young, J. D., Shastri, A. A., Stephanopoulos, G. & Morgan, J. A. Mapping photoautotrophic metabolism with isotopically nonstationary 13C flux analysis. Metab. Eng. 14, 185–185 (2012).
Xiong, W. et al. The plasticity of cyanobacterial metabolism supports direct CO2 conversion to ethylene. Nature Plants 1, 15053 (2015).
Carrieri, D., Paddock, T., Maness, P. C., Seibert, M. & Yu, J. P. Photo-catalytic conversion of carbon dioxide to organic acids by a recombinant cyanobacterium incapable of glycogen storage. Energ. Environ. Sci. 5, 9457–9461 (2012).
Grundel, M., Scheunemann, R., Lockau, W. & Zilliges, Y. Impaired glycogen synthesis causes metabolic overflow reactions and affects stress responses in the cyanobacterium Synechocystis sp PCC 6803. Microbiology 158, 3032–3043 (2012).
McNeely, K. et al. Synechococcus sp. strain PCC 7002 nifJ mutant lacking pyruvate:ferredoxin oxidoreductase. Appl. Environ. Microbiol. 77, 2435–2444 (2011).
Zhou, J., Zhang, H. F., Zhang, Y. P., Li, Y. & Ma, Y. H. Designing and creating a modularized synthetic pathway in cyanobacterium Synechocystis enables production of acetone from carbon dioxide. Metab. Eng. 14, 394–400 (2012).
Fandi, K. G., Ghazali, H. M., Yazid, A. M. & Raha, A. R. Purification and N-terminal amino acid sequence of fructose-6-phosphate phosphoketolase from Bifidobacterium longum BB536. Lett. Appl. Microbiol. 32, 235–239 (2001).
Meile, L., Rohr, L. M., Geissman, T. A., Herensperger, M. & Teuber, M. Characterization of the D-xylulose 5-phosphate/D-fructose 6-phosphate phosphoketolase gene (xfp) from Bifidobacterium lactis. J. Bacteriol. 183, 2929–2936 (2001).
Amaya-Delgado, L., Hidalgo-Lara, M. E. & Montes-Horcasitas, M. C. Characterization of the D-xylulose 5-phosphate/D-fructose 6-phosphate phosphoketolase gene, xpkL, from Cellulomonas flavigena. J. Biotechnol. 118, S134–S134 (2005).
Liu, L. X. et al. Phosphoketolase pathway for xylose catabolism in Clostridium acetobutylicum revealed by 13C metabolic flux analysis. J. Bacteriol. 194, 5413–5422 (2012).
Posthuma, C. C. et al. Expression of the xylulose 5-phosphate phosphoketolase gene, xpkA, from Lactobacillus pentosus MD363 is induced by sugars that are fermented via the phosphoketolase pathway and is repressed by glucose mediated by CcpA and the mannose phosphoenolpyruvate phosphotransferase system. Appl. Environ. Microbiol. 68, 831–837 (2002).
Duan, Z. B., Shang, Y. F., Gao, Q., Zheng, P. & Wang, C. S. A phosphoketolase Mpk1 of bacterial origin is adaptively required for full virulence in the insect-pathogenic fungus Metarhizium anisopliae. Environ. Microbiol. 11, 2351–2360 (2009).
Sarkar, P. & Roy, A. Molecular cloning, characterization and expression of a gene encoding phosphoketolase from Termitomyces clypeatus. Biochem. Biophys. Res. Commun. 447, 621–625 (2014).
Moriyama, T., Tajima, N., Sekine, K. & Sato, N. Characterization of three putative xylulose 5-phosphate/fructose 6-phosphate phosphoketolases in the cyanobacterium Anabaena sp. PCC 7120. Biosci. Biotechnol. Biochem. 79, 767–774 (2015).
Suzuki, R. et al. Crystal structures of phosphoketolase thiamine diphosphate-dependent dehydration mechanism. J. Biol. Chem. 285, 34279–34287 (2010).
You, L., Berla, B., He, L., Pakrasi, B. H. & Tang, Y. J. 13C-MFA delineates the photomixotrophic metabolism of Synechocystis sp. PCC 6803 under light- and carbon-sufficient conditions. Biotechnol. J. 9, 684–692 (2014).
Nakamura, Y., Kaneko, T., Hirosawa, M., Miyajima, N. & Tabata, S. CyanoBase, a WWW database containing the complete nucleotide sequence of the genome of Synechocystis sp. strain PCC6803. Nucleic Acids Res. 26, 63–67 (1998).
Kucho, K. et al. Global analysis of circadian expression in the cyanobacterium Synechocystis sp strain PCC 6803. J. Bacteriol. 187, 2190–2199 (2005).
Battchikova, N. et al. Dynamic changes in the proteome of Synechocystis 6803 in response to CO2 limitation revealed by quantitative proteomics. J. Proteome Res. 9, 5896–5912 (2010).
Kaczmarzyk, D. & Fulda, M. Fatty acid activation in cyanobacteria mediated by acyl-acyl carrier protein synthetase enables fatty acid recycling. Plant Physiol. 152, 1598–1610 (2010).
Wu, G., Bao, T., Shen, Z. & Wu, Q. Sodium acetate stimulates PHB biosynthesis in Synechocystis sp. PCC 6803. Tsinghua Sci. Technol. 7, 435–438 (2002).
Bogorad, I. W., Lin, T. S. & Liao, J. C. Synthetic non-oxidative glycolysis enables complete carbon conservation. Nature 502, 693–697 (2013).
Kocharin, K., Siewers, V. & Nielsen, J. Improved polyhydroxybutyrate production by Saccharomyces cerevisiae through the use of the phosphoketolase pathway. Biotechnol. Bioeng. 110, 2216–2224 (2013).
Antoniewicz, M. R., Kelleher, J. K. & Stephanopoulos, G. Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab. Eng. 9, 68–86 (2007).
Acknowledgements
This work was supported by National Renewable Energy Laboratory Director's Postdoc Fellowship (to W.X.), and by the US Department of Energy (DOE), Office of Science, Basic Energy Science (to M.G., M.C., J.Y.). The latter funded in part the conception and execution of the work as well as preparation of the manuscript. It was also supported in part by the DOE Office of Energy Efficiency and Renewable Energy, Fuel Cell Technologies Office (to P.C.M.), BioEnergy Technologies Office (to E.G.), Science Undergraduate Laboratory Internship program (to S.R.), and a Dragon-Gate grant (to T.C.L.) from Ministry of Science and Technology in Taiwan. The authors acknowledge M. Seibert from NREL and A. Grossman from the Carnegie Institution for Science for helpful discussion. The software Metran was developed and kindly provided by Maciek R. Antoniewicz from the University of Delaware. The US Government retains and the publisher, by accepting the article for publication, acknowledges that the US Government retains a nonexclusive, paid up, irrevocable, worldwide license to publish or reproduce the published form of this work, or allow others to do so, for US Government purposes.
Author information
Authors and Affiliations
Contributions
W.X., P.C.M., M.G. and J.Y. planned the project; W.X., T.C.L., E.G., M.C. and S.R. performed the experiments; W.X., E.G. and J.Y. analysed the data and drafted the manuscript; all authors edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Xiong, W., Lee, TC., Rommelfanger, S. et al. Phosphoketolase pathway contributes to carbon metabolism in cyanobacteria. Nature Plants 2, 15187 (2016). https://doi.org/10.1038/nplants.2015.187
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2015.187
This article is cited by
-
Switching off the lights to illuminate a photosynthetic brake in cyanobacteria
Nature Metabolism (2023)
-
A parallel glycolysis provides a selective advantage through rapid growth acceleration
Nature Chemical Biology (2023)
-
Optimal energy and redox metabolism in the cyanobacterium Synechocystis sp. PCC 6803
npj Systems Biology and Applications (2023)
-
GC/MS-based 13C metabolic flux analysis resolves the parallel and cyclic photomixotrophic metabolism of Synechocystis sp. PCC 6803 and selected deletion mutants including the Entner-Doudoroff and phosphoketolase pathways
Microbial Cell Factories (2022)
-
Engineering microbial pathways for production of bio-based chemicals from lignocellulosic sugars: current status and perspectives
Biotechnology for Biofuels (2020)