Abstract
Background:
The waterflea Daphnia is an interesting candidate for bioregenerative life support systems (BLSS). These animals are particularly promising because of their central role in the limnic food web and its mode of reproduction. However, the response of Daphnia to altered gravity conditions has to be investigated, especially on the molecular level, to evaluate the suitability of Daphnia for BLSS in space.
Methods:
In this study, we applied a proteomic approach to identify key proteins and pathways involved in the response of Daphnia to simulated microgravity generated by a two-dimensional (2D) clinostat. We analyzed five biological replicates using 2D-difference gel electrophoresis proteomic analysis.
Results:
We identified 109 protein spots differing in intensity (P<0.05). Substantial fractions of these proteins are involved in actin microfilament organization, indicating the disruption of cytoskeletal structures during clinorotation. Furthermore, proteins involved in protein folding were identified, suggesting altered gravity induced breakdown of protein structures in general. In addition, simulated microgravity increased the abundance of energy metabolism-related proteins, indicating an enhanced energy demand of Daphnia.
Conclusions:
The affected biological processes were also described in other studies using different organisms and systems either aiming to simulate microgravity conditions or providing real microgravity conditions. Moreover, most of the Daphnia protein sequences are well-conserved throughout taxa, indicating that the response to altered gravity conditions in Daphnia follows a general concept. Data are available via ProteomeXchange with identifier PXD002096.
Similar content being viewed by others
Introduction
Since the first moon landing in 1969, technologies have advanced and the urge to further explore space has not diminished. At the moment, the ultimate goal of manned space missions is the exploration of Mars. Yet manned missions face several key issues that have to be solved, not only concerning human health,1 but also the provision with essential supplies, e.g., food, water, oxygen.
A solution for this supply problem, especially for long duration missions, is the development of a bioregenerative life support system (BLSS), which minimizes reliability on delivered supplies and enhances autochtonous production. Up to now, such systems have only been installed as modules on the Mir and ISS space stations.2 Since then, new components were added to the life support system, e.g., waste water recovery, forming a so-called ecological control and life support system.3 The functions that have to be fulfilled by this system are the regeneration of atmosphere, purification of water, waste processing, food production, and food processing. The organisms involved in these systems include bacteria and fungi for the decomposition of organic waste and excrements, as well as unicellular microalgae, which produce the oxygen for astronauts, but also comprise higher organisms such as vegetables or fish to provide the astronauts with food.4
The waterflea Daphnia (Crustacea) might be a candidate in aquatic modules of such a BLSS for several reasons: (i) Daphnia occupies a central role in limnic food webs by being a primary consumer, hence serving as a link between oxygen producing, autotrophic producers such as algae and secondary consumers, such as planktivorous fish.5 Fish, in respect, serve as an animal protein source for the human crew. Positive side effects of this constellation are that no additional fish food has to be transported, as well as that the growth of algae populations is controlled. (ii) Daphnia reproduces by the mode of cyclic parthenogenesis, thus enabling Daphnia to reproduce asexually in favorable environmental conditions and sexually in unfavorable ones, which leads to the formation of dormant eggs that are encased in a protective structure, the so called ephippium. Those resting eggs could be used to restart the BLSS in case of a system collapse and it was already shown that dormant eggs of Daphnia are able to hatch living neonates after long-term exposure to the space environment on the ISS.6 Combining these modes of reproduction with the short generation time and the high number of offspring per clutch, a high bio mass production can be guaranteed.7
Daphnia does not only serve as a model organism in the fields of ecology, evolution, and ecotoxicology,8 but also in ecological genomics.9 Here, especially the improved availability of genomic resources10 facilitates untargeted holistic approaches, such as transcriptomics11 or proteomics,12,13 which may reveal unpredicted key players underlying biological traits.
Long-term spaceflight is known to affect human physiology leading to bone demineralization, skeletal muscle atrophy, and immune system suppression.1 Furthermore, microgravity-induced responses were described in a variety of biological systems reaching from alterations of cytoskeletal formation in cells14 to altered plant forms in Arabidopsis.15 Daphnia has already been part of missions to space stations, demonstrating that resting eggs were viable even after exposure to outer space for >1 year.6 Some animals survived up to 4 months in space but showed alterations in swimming behavior with an unusual high proportion of looping movements.16 However, more information on the influence of microgravity on Daphnia is needed to decide on the suitability of these animals for BLSS in long-duration missions. Studies at the molecular level are especially interesting, as they may elucidate additional biological processes not detectable at the morphological or physiological level.
To determine the effect of long-term exposure to weightlessness, high technical complexity and financial investment is needed. Most facilities providing free fall conditions, like parabolic flights or drop-tower experiments can only deliver short duration of weightlessness. Yet a cost-effective ground-based method is the use of a two-dimensional (2D)-clinostat,17 however, carefully considering the operational mode and limitation of the simulation.
In this study, we investigated the effect of simulated microgravity on Daphnia at the protein level, as proteins are the main effectors of biological functions in an organism. We exposed Daphnia to simulated microgravity using a 2D-clinostat and subsequently performed a proteomic approach to study quantitative changes in the proteome of animals exposed to altered gravity conditions compared with a control group.
Materials and methods
Animal husbandry
To investigate the effect of simulated microgravity on the proteome of Daphnia magna the laboratory cultivated genotype K34J was used, which originated from a fishpond near Munich, Germany. The animals were kept in a density of 5–12 adult animals per 1.5-l jar filled with semi-artificial medium18 in an illuminated climate chamber (Binder KBWF 240, Binder GmbH, Tuttlingen, Germany) at 21±1 °C and a photoperiod of 12 h (L18W 865 Cool Daylight, Osram, Munich, Germany). Animals were fed every second day with the unicellular algae Scenedesmus obliquus, thereby reaching a carbon concentration of 1.5 mg/l.
Simulated microgravity—clinostat experiment
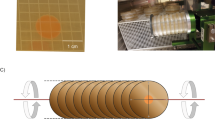
To simulate microgravity, we used a cuvette 2D-clinostat, designed and provided by the German Aerospace Center (Deutsches Zentrum für Luft- und Raumfahrt, DLR) in Cologne, Germany (Figure 1).
The clinorotation principle is based on the fast rotation around a small diameter thereby preventing physical sedimentation.19 As a consequence of clinorotation, the composition of the D. magna proteome was investigated in comparison to the proteome under normal gravity conditions, which means in a static 1 g control. For the experiment, D. magna of 2±0.2 mm size were used. Body size was measured with a Leica MS5 stereomicroscope (Leica Microsystems, Wetzlar, Germany). The clinostat was loaded with 10 1-ml serological BD-Falcon pipettes (BD Biosciences, Heidelberg, Germany). Each pipette contained 1 ml of medium enriched with algae (carbon concentration: 1.5 mg/l) and five randomly selected D. magna. Animals were able to move freely in the cuvette. Alternately one pipette was mounted on the clinostat and the next one was put next to the clinostat as a control, therefore the control pipettes were exposed to the same vibrations as the clinorotated pipettes. This process was repeated until 10 pipettes of each treatment were arranged within the experimental setup.
The rotation speed of the clinostat was set at 60 r.p.m., with a residual gravity of ~0.008g.20 The duration of the experiment was set to 60 min at a room temperature of 20 °C. Longer exposure times of several days, as implemented for plants,17 would not be possible without creating food limitations for Daphnia, especially as a higher starting amount of algae may have harmful effects on the animals and adding of additional algae is not feasible during clinorotation because of turbulence generation. However, as an exposure time of 60 min was sufficient to show effects in other studies, e.g., study by Eiermann et al.,21 and the algae concentration was sufficient for this amount of time, we chose this duration for our clinorotation experiment. After 60 min, the pipettes were emptied during rotation by tilting the device into cryo tubes and excessive water was immediately removed and cryo tubes were snap frozen in liquid nitrogen. The process of water removal and freezing was performed in less than 5 s. A total number of 10 runs was performed, each consisting of 10 serological cuvettes and therefore 50 animals of each treatment.
Sample preparation
To generate samples for proteomic analysis, the frozen biological samples were pulverized in a mortar containing liquid nitrogen to prevent thawing. The resulting powder was solubilised in 330 μl lysis buffer (2 mol/l Thiourea, 6 mol/l Urea, 4% CHAPS, 1 cOmplete ULTRA Tablets Mini (Roche, Penzberg, Germany) per 5 ml buffer). Afterwards, each sample was centrifuged using a QIA Shredder Mini Spin Column (Qiagen, Hilden, Germany) for 3 min at 16,100 r.c.f. to get rid of debris. Proteins were precipitated using 30% trichloroacetic acid for 20 min on ice to inhibit proteolytic activity.22 Subsequently, samples were centrifuged for 10 min at 16,100 r.c.f., the supernatant was discarded and the protein pellet was washed three times with cold acetone. The pellet was dried and resolved in lysis buffer. The pH of the solution was adjusted to 8 by adding 50 mM NaOH. Protein concentration was determined by Bradford Assay (Coomassie Plus (Bradford) Assay Reagent, Thermo Scientific, Braunschweig, Germany) according to the manufacturer’s instructions. To reach sufficient protein concentrations for 2D-difference gel electrophoresis (DIGE), two clinorotation runs per group were pooled leading to five biological replicates.
2D-DIGE
2D-DIGE method was conducted following the general procedure described in the study by Otte et al.13 Briefly, 50 μg protein per biological replicate was labeled with 2D-DIGE Cy3 Dye for control and Cy5 Dye for the clinorotated group following the protocol of the manufacturer (GE Healthcare Life Sciences, Munich, Germany). An internal pooled standard was prepared by pooling 25 μg protein of all biological replicates and labeling of 300 μg with 2D-DIGE Cy2 Dye. For each 2D-DIGE gel, 50 μg of one Cy3-labeled control replicate, 50 μg of one Cy5-labeled clinorotated replicate, and 50 μg of Cy2-labeled internal pooled standard were combined.
For first dimension separation, 24-cm gel strips (DryStrips pH 4–7, GE Healthcare) and an IPGPhore (Pharmacia Biotech, Uppsala, Sweden) were used. For second dimension separation, the gel strips were fixed on top of lab cast gels and electrophoresis was performed using an ETTANDaltsix electrophoresis unit (GE Healthcare Life Sciences). During the whole 2D-DIGE procedure, all five biological replicates were processed in parallel.
Imaging and quantitative analysis
Immediately after electrophoresis, gels were scanned using a Typhoon 9400 fluorescence scanner (GE Healthcare Life Sciences) with parameter settings recommended by the manufacturers for 2D-DIGE experiments. Image analysis and relative quantification were performed with DeCyder 2D Software version v7.0 (GE Healthcare Life Sciences). Coordinates of corresponding spots differing significantly in their intensity (P⩽0.05 after false discovery rate correction, ratio ⩾|2|) were used to generate a pick list for further processing.
Excision of spots and tryptic hydrolysis
Gels were stained overnight with colloidal Coomassie staining solution (Carl Roth GmbH, Karlsruhe, Germany) and then destained using 25% methanol. Further processing of proteins spots was performed according to in the study by Otte et al.13 In summary, spots of interest were cut out of the gel and digested with trypsin (Sequencing Grade–Modified Trypsin, Promega, Mannheim, Germany) to generate peptides for protein identification.
LC-MS/MS analysis
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed on a multi-dimensional LC system (Ettan MDLC, GE Healthcare Life Sciences) coupled to a LTQ mass spectrometer (Thermo Scientific, Braunschweig, Germany). Further settings were the same as described in the study by Otte et al.13
Bioinformatic processing
For protein identification, MS/MS data were searched with Mascot Version: 2.3.00 (Matrix Science, London, United Kingdom). As database, pre-released gene-predictions of D. magna (V2.4 effective May 2012) were used. These sequence data were produced by The Center for Genomics and Bioinformatics at Indiana University and distributed via wFleaBase in collaboration with the Daphnia Genomics Consortium (http://daphnia.cgb.indiana.edu). Further data processing was done as described in the study by Otte et al.13 Protein spots having multiple protein identifications were not included in the final data set. To get further information on similar proteins, all significant protein sequences were blastp searched against NCBI non-redundant (nr) and Swiss-Prot databases using local standalone blast.23 The NCBI nr database combines non-redundant protein sequences of several sources, including translations from annotated coding regions in GenBank, RefSeq, and TPA, as well as records from PIR, PRF, and PDB, whereas the Swiss-Prot database is manually annotated and therefore contains less but more reliable data. Thus, recent data of genomic studies on several organisms are found in nr database, whereas Swiss-Prot consist mainly of well-annotated protein information of a few well-studied model organisms.
Preliminary annotation data were received from http://server7.wfleabase.org/genome/Daphnia_magna/ (V2.4 effective May 2012). Protein-associated gene ontology (GO) terms were tested for enrichment using customized standalone EASE24 (Benjamini-corrected P value<0.05). Enriched terms were tested for redundant terms and semantic similarities using the online-tool REViGO25 and visualized as treemap graphs using R.26
In addition, protein sequences were also processed using the software Blast2go,27 which uses results of NCBI blast search to map sequences directly to GO terms. These GO terms were used as additional information.
Results
We analyzed the effects of altered gravity conditions on D. magna at the protein level by exposing animals for 1 h to clinorotation followed by a proteomic 2D-DIGE approach. We studied five clinorotated and five control replicates. We generated 5 2D-DIGE gels, which showed reproducible spot map patterns (Supplementary Data S2), and were able to match and quantify 1,211 protein spots in at least 4 of these 5 spot maps. Unsupervised hierarchical clustering of spot intensity data present in all spot maps showed a good clustering according to treatment (Figure 2).
Example 2D-DIGE gel image. Here, protein spots which were significantly different between treatments and were identified using LC-MS/MS were marked. Red labeled spots were less abundant in the clinorotated treatment, whereas white labeled spots were more abundant in the clinorotated treatment. LC-MS/MS, liquid chromatography-tandem mass spectrometry; 2D-DIGE, two-dimensional difference gel electrophoresis.
About 109 of these protein spots showed significantly different intensity signals between the control and the clinorotated treatment (P⩽0.05, ratio ⩾|2|) and were identified using LC-MS/MS (Figure 3 and Table 1). Of these protein spots, 30 were less abundant, whereas 79 protein spots were more abundant in the clinorotated treatment.
As information on protein function of D. magna proteins is scarce so far, we performed a blastp search against NCBI nr and Swiss-Prot database to look for similar and possibly better characterized proteins in other species. Here, we used a strict BLAST bit score threshold of ⩾244, which ensures accurate prediction of protein function similarity.28 Interestingly, nearly all hits in the nr database refer to proteins of D. pulex. In contrast to D. magna, the D. pulex genome is published10 and therefore completely represented in this database. Furthermore, a majority of proteins had a similar blast hit in the Swiss-Prot database, indicating the presence of similar proteins in well-studied model organisms and therefore a good conservation of these proteins within organismal taxa (Supplementary Data S1).
Enrichment analysis of GO terms yielded five overrepresented terms in biological process and molecular function database, namely protein folding, unfolded protein binding, actin binding, ATP binding, and glycolytic process (Figure 4 and Table 2). Furthermore, nine protein spots were identified as Daphnia hemoglobins.
About 27 protein spots were identified as proteins connected to actin binding and they were either involved in muscular structures or the cytoskeleton. Beyond actin itself, we identified myosin, α-actinin, filamin-A, gelsolin, and advillin. Some of these proteins are present in multiple spots with isoelectric point and molecular weight shifts, and varying abundances, indicating post-translational modifications (PTMs). Most spots identified as actin, myosin, α-actinin, and filamin-a were less abundant, whereas advillin and gelsolin were identified in spots to be more abundant after clinorotation.
Furthermore, 20 protein spots were identified as proteins involved in protein folding. Here, heat shock proteins and other chaperones like endoplasmin, a protein disulfide-isomerase, and different subunits of T-complex protein 1 were found. All spots except one had a higher abundance in the animals exposed to clinorotation.
In addition, 17 protein spots were connected to different metabolic pathways involved in energy generation. Proteins were involved in glycolysis, a GO term which was also found to be overrepresented in our data set, in the tricarboxylic acid cycle, the respiratory chain, or pentose phosphate pathway. All proteins except fructose-bisphosphate-aldolase were more abundant in Daphnia exposed to simulated microgravity.
For a summary of identified proteins see Table 3, for details on protein spots see Supplementary Data S1.
Discussion
To analyze the effects of altered gravity conditions on the waterflea D. magna, we exposed animals to fast clinorotation, an established method to simulate microgravity conditions on ground. Nevertheless one has to keep in mind, that this kind of simulation approach has to be verified under real microgravity conditions.17 The clinorotated animals were compared with a control group using a proteomic 2D-DIGE approach (five biological replicates). As a general result of this study, proteins involved in actin microfilament organization were less abundant in clinorotated animals, whereas proteins connected to protein folding and energy metabolism were more abundant.
Exposure to simulated microgravity may disrupt actin microfilament organization in Daphnia
Around 25% of the significantly altered and identified protein spots in our data set were related to muscular structures or the cytoskeleton. All of these proteins were annotated with the GO term actin binding, which was also found to be significantly overrepresented (Figure 4).
We identified the structural protein actin in 12 spots, showing isoelectric point and molecular weight shifts on the 2D-gel. Two different kinds of spots were observed, indicating two groups of PTMs. The first group consists of four spots which were more abundant in the clinorotated treatment and have a pI near the theoretical value but an increased molecular mass. The second group of eight spots is less abundant in the clinorotated treatment and has a decreased pI but the molecular weight is near the theoretical value or below (Supplementary Data S1). These spots, which were located differently on the gel showing different abundance ratios, may indicate changes of actin PTMs. These changes may be related to alterations of actin microfilament organization, as PTMs are known to modulate structure and function of actin.29
The effect of clinorotation on actin filaments in our study is further emphasized by abundance alterations of actin-related proteins. Here, not only the actin-filament-associated motor protein myosin but also the actin-binding proteins α-actinin and filamin-A were less abundant in animals exposed to simulated microgravity. Filamin is known to be involved in the recruitment of actin filaments in Drosophila30 and its decreased abundance may therefore indicate cytoskeletal disorganization. α-actinin is a cross-linker of actin filaments in muscle and non-muscle cells. Drosophila knock-down mutants are known to suffer from muscle weakness and atrophy,31 which may also be the case in D. magna exposed to microgravity. Furthermore, α-actinin was also found to be less abundant in human neuroblastoma cells exposed to simulated microgravity.32 In contrast to these proteins, the actin-binding proteins advillin and gelsolin were more abundant in clinorotated Daphnia. Both proteins are members of the gelsolin/villin family, which are involved in the regulation of actin polymer organization33 and are also able to sever actin filaments. Therefore, it can be stated that our findings on actin-related proteins provide strong evidence for a disruption of actin microfilament organization in D. magna exposed to simulated microgravity, at least in the time frame of 60-min exposure.
Alterations of the cytoskeleton as a result of different gravity conditions are a phenomenon described in many studies analyzing various mammalian cell lines either exposed to real or simulated microgravity.14 In several studies, the disorganization of the actin cytoskeleton was observed in human monocytes34 and human neuroblastoma cells32 using immunohistochemistry and studying cell morphology. Furthermore, changes of cytoskeletal element gene expression and morphology in human cells were detected on a short time-scale after only 2 s in real microgravity during parabolic flights.35,36 In addition, actin protein abundance decreased after 12 days of spaceflight in Arabidopsis.37
Large-scale effects of microgravity on the muscular system of humans, mice, and rats are also well-studied,38 leading to atrophy and reduced functional capacity of the muscles. However, the source and function of actin found in our study, either cytoplasmic or muscular, is hard to predict without further experiments. Röper et al.39 showed that muscle-specific actin is incorporated into cytoplasmic structures, and cytoskeletal actin is incorporated into muscles for all actin paralogues of Drosophila Melanogaster. Therefore, it is not possible to deduce actin function solely from its protein sequence.
Chaperones are involved in the stress response of Daphnia to simulated microgravity
Another substantial fraction of protein spots were associated with protein binding, a GO term found overrepresented in the enrichment analysis (Figure 4). In this group, several molecular chaperones were identified with nearly all proteins being more abundant in the clinorotated treatment.
Heat shock proteins and other chaperones facilitate protein folding, unfolding, and transportation40 playing an important role in both normal cellular homeostasis and stress response.41 In Daphnia, they were found to be involved in responses to several stressors, e.g., temperature changes,11 presence of a predator,13,42 exposure to the drug diclofenac,43 and exposure to copper.44
Activation of the heat shock system was also observed as response to microgravity in other systems, leading to increased gene expression or higher abundance of heat shock proteins. This was reported for plant cells exposed to simulated microgravity and microgravity during spaceflight,45 for animal cells exposed to simulated microgravity32 and also for Drosophila during spaceflight.46 Our data clearly indicate that in Daphnia, the heat shock system possibly reacts to stress-dependent changes in cell, tissue, or organ structures caused by altered mechanical (gravitational) forces.47 Therefore, proteins related to protein folding seem to be involved in the response to microgravity.46
In addition to heat shock proteins, we identified subunits of the chaperonin containing T-complex protein, also known as CCT, in several spots, which were all more abundant in the clinorotated treatment. CCT is known to contribute to the folding of a distinct subset of cellular proteins including cytoskeletal proteins like actin and myosin.48 Moreover, protein disulfide-isomerase was more abundant in the clinorotated animals. This protein is an essential folding catalyst and chaperone located in the endoplasmatic reticulum, which introduces disulfide bonds into proteins and catalyses the rearrangement of incorrect disulfides.49
Therefore, the high abundance of proteins related to protein folding in Daphnia exposed to clinorotation is a strong indicator for a microgravity-induced breakdown of protein structures in general.
Clinorotation leads to an increased energy demand in Daphnia
Further molecular consequences of simulated microgravity on Daphnia is seen in the field of energy related proteins. Here, proteins associated with various energy pathways were altered in their abundance, most of them more abundant in the clinorotated treatment.
We found proteins involved in glycolysis (enolase, phosphoglycerate kinase, and fructose-bisphosphate aldolase), the TCA cycle (malate dehydrogenase), and in the respiratory chain (NADH-ubiquinone oxidoreductase and ATP synthase subunit β). Most of these proteins were more abundant in the clinorotated treatment, indicating an enhanced energy metabolism in simulated microgravity. Furthermore, the enhanced abundance of glycogen phosphorylase is also an indicator of increased energy consumption in clinorotated animals, as it catalyses the degradation of glycogen to provide an increased amount of glucose.
In contrast to the other glycolytic proteins, fructose-bisphosphate aldolase was less abundant in clinorotated animals. However, this protein is also involved in processes apart from glycolysis, e.g., pentose phosphate pathway, and fructose and mannose metabolism, which may also be affected by the stressful condition of altered gravity. Similarly, transketolase, is also involved in the pentose phosphate pathway and showed only a slightly higher abundance in clinorotated Daphnia.
Interestingly, proteins related to energy metabolism were also found to be affected by microgravity in other systems. In Arabidopsis thaliana grown for 12 days on the ISS, a lower abundance of these proteins was observed.37 In contrast, exposure to simulated microgravity for several hours using a clinostat led to an increased abundance of carbohydrate metabolism proteins in Arabidopsis callus50 and root cells.51 The differences that were found in the abovementioned studies might be based on different exposure times (short-term versus long term) or different exposure methods (simulated versus real microgravity), which may influence the response to microgravity.17 However, indications for an increased energy metabolism found in our study correlated well with the similar clinorotation experiments in A. thaliana.50,51 Changes in protein abundance related to energy metabolism were also observed in Pseudomonas aeruginosa, an opportunistic pathogen, when exposed to microgravity. Here, proteins related to glycolysis were of higher abundance in ground control treatments when compared with P. aeruginosa exposed to microgravity.52 Furthermore, alterations in the expression of proteins related to energy metabolism were also reported in the bacterial pathogen Salomonella in spaceflight environment.53,54
Another indicator for an increased energy demand in Daphnia in our study is the altered abundance of hemoglobin. Daphnia, is known to increase its hemoglobin concentration in response to environmental hypoxia as well as to temperature increase.55 Here, elevated water temperature has two effects, a decrease of oxygen partial pressure due to decreased oxygen solubility and an increase of metabolic rate in the ectotherm animal.
Daphnia hemoglobin genes are located in a tandem-duplicated gene cluster, which contributes to the varying composition of the protein. Both, the abundance of different Hb subunits, as well as their post translational modifications change in an oxygen-dependant manner, most probably optimizing oxygen affinity of hemoglobin according to oxygen concentration.56 The same may be true for our study, explaining the occurrence of both, more and less abundant hemoglobin spots in the clinorotated treatment with pI and mass shifts.
As average hemoglobin abundance is higher in clinorotated animals, oxygen consumption may be increased. Therefore, hemoglobin abundance is most probably not primarily affected by microgravity, but clinorotation may lead to a stress response in Daphnia, resulting in higher energy demand and therefore increased oxygen consumption. As the amount of available oxygen in the cuvette is limited due to the experimental device, this increased oxygen consumption may lead to hypoxic conditions induced by the experimental device rather than microgravity. Animals in the control cuvette did not show an increase in hemoglobin abundance although oxygen concentration in the cuvette was similar, which indicates higher oxygen consumption of clinorotated animals. However, it cannot be excluded that the increased energy demand found in our study is related to altered swimming behavior of clinorotated Daphnia, as it is known that Daphnia in space shows an increased amount of looping/summersaulting movements.16
Conclusion
Using a proteomic approach, we were able to identify molecular key processes affected by clinorotation. Assuming that this simulation approach is suitable for Daphnia, which has to be verified in space, our results indicate impacts of microgravity on the parameters of investigations. We found strong indicators of actin cytoskeleton disruption and breakdown of protein structures in general and an increase of energy demands. These results are in agreement with results obtained from other organisms and microgravity systems. Interestingly, most of the proteins found to be affected are well-conserved throughout taxa.
Our proteomic approach led to interesting insights into the responses to altered gravity conditions. These results not only resemble important starting points for further Daphnia space research concerning life support systems but also increase the knowledge on the influence of gravity on biochemical processes. In addition, our data strongly suggest that a lack of gravity affects similar molecular processes in a variety of organisms.
References
Guéguinou N, Huin-Schohn C, Bascove M, Bueb JL, Tschirhart E, Legrand-Frossi C et al. Could spaceflight-associated immune system weakening preclude the expansion of human presence beyond Earth’s orbit? J Leukoc Biol 2009; 86: 1027–1038.
Sychev VN, Levinskikh Ma, Shepelev YY . The biological component of the life support system for a martian expedition. Adv Space Res 2003; 31: 1693–1698.
Wieland PO . Living together in space: the design and operation of the life support systems on the International Space Station. National Aeronautics and Space Administration Marshall Space Flight Center MSFC, Huntsville, AL, USA, 1998. Reportno. NASA/TM—98–206956/VOL1.
Schwartzkopf S . Design of a controlled ecological life support system. BioScience 1992; 42: 526–535.
Lampert W . Daphnia: model herbivore, predator and prey. Pol J Ecol 2006; 54: 607–620.
Novikova N, Gusev O, Polikarpov N, Deshevaya E, Levinskikh M, Alekseev V et al. Survival of dormant organisms after long-term exposure to the space environment. Acta Astronaut 2011; 68: 1574–1580.
Sakwinska O . Plasticity of Daphnia magna life history traits in response to temperature and information about a predator. Freshw Biol 1998; 39: 681–687.
Stollewerk A . The water flea Daphnia-a’new’model system for ecology and evolution? J Biol 2010; 9: 2–5.
Miner BE, De Meester L, Pfrender ME, Lampert W, Hairston NG . Linking genes to communities and ecosystems: Daphnia as an ecogenomic model. Proc Biol Sci 2012; 279: 1873–1882.
Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH et al. The ecoresponsive genome of Daphnia pulex . Science 2011; 331: 555–561.
Yampolsky LY, Zeng E, Lopez J, Williams PJ, Dick KB, Colbourne JK et al. Functional genomics of acclimation and adaptation in response to thermal stress in Daphnia . BMC genomics 2014; 15.
Fröhlich T, Arnold GJ, Fritsch R, Mayr T, Laforsch C . LC-MS/MS-based proteome profiling in Daphnia pulex and Daphnia longicephala: the Daphnia pulex genome database as a key for high throughput proteomics in Daphnia . BMC Genomics 2009; 10.
Otte K, Fröhlich T, Arnold G, Laforsch C . Proteomic analysis of Daphnia magna hints at molecular pathways involved in defensive plastic responses. BMC Genomics 2014; 15.
Vorselen D, Roos WH, MacKintosh FC, Wuite GJL . Van Loon JJWa. The role of the cytoskeleton in sensing changes in gravity by nonspecialized cells. FASEB J 2014; 28: 536–547.
Link BM, Busse JS, Stankovic B . Seed-to-seed-to-seed growth and development of Arabidopsis in microgravity. Astrobiology 2014; 14: 866–875.
Ijiri K, Mizuno R, Narita T, Ohmura T, Ishikawa Y, Yamashita M et al. Behavior and reproduction of invertebrate animals during and after a long-term microgravity: space experiments using an autonomous biological system(ABS). Biol Sci Space 1998; 12: 377–388.
Herranz R, Anken R, Boonstra J, Braun M, Christianen PCM, de Geest M et al. Ground-based facilities for simulation of microgravity: organism-specific recommendations for their use, and recommended terminology. Astrobiology 2013; 13: 1–17.
Rabus M, Laforsch C . Growing large and bulky in the presence of the enemy: Daphnia magna gradually switches the mode of inducible morphological defences. Funct Ecol 2011; 25: 1137–1143.
Herranz R, Manzano AI, van Loon JJWa, Christianen PCM, Medina FJ . Proteomic signature of Arabidopsis cell cultures exposed to magnetically induced hyper- and microgravity environments. Astrobiology 2013; 13: 217–224.
Brungs S, Hauslage J, Hilbig R, Hemmersbach R, Anken R . Effects of simulated weightlessness on fish otolith growth: clinostat versus rotating-wall vessel. Adv Space Res 2011; 48: 792–798.
Eiermann P, Kopp S, Hauslage J, Hemmersbach R, Gerzer R, Ivanova K . Adaptation of a 2-D clinostat for simulated microgravity experiments with adherent cells. Micrograv Sci Technol 2013; 25: 153–159.
Zeis B, Lamkemeyer T, Paul RJ, Nunes F, Schwerin S, Koch M et al. Acclimatory responses of the Daphnia pulex proteome to environmental changes. I. Chronic exposure to hypoxia affects the oxygen transport system and carbohydrate metabolism. BMC Physiol 2009; 9.
Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S et al. The NCBI BioSystems database. Nucleic Acids Res 2010; 38: D492–D496.
Hosack DA, Dennis GJ, Sherman BT, Lane HC, Lempicki RA . Identifying biological themes within lists of genes with EASE. Genome Biol 2003; 4: R70.
Supek F, Bošnjak M, Škunca N, Šmuc T . REVIGO summarizes and visualizes long lists of gene ontology terms. PloS One 2011; 6: e21800.
R Core Team (2015). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria; https://www.R-project.org/.
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M . Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005; 21: 3674–3676.
Louie B, Higdon R, Kolker E . A statistical model of protein sequence similarity and function similarity reveals overly-specific function predictions. PloS One 2009; 4: e7546.
Terman JR, Kashina A . Post-translational modification and regulation of actin. Curr Opin Cell Biol 2013; 25: 1–9.
Sokol N, Cooley L . Drosophila Filamin encoded by the cheerio locus is a component of ovarian ring canals. Curr Biol 1999; 9: 1221–1230.
Roulier E, Fyrberg C, Fyrberg E . Perturbations of Drosophila alpha-actinin caus muscle paralysis, weakness, and atrophy but do not confer obvious nonmuscle phenotypes. J Cell Biol 1992; 116: 911–922.
Zhang Y, Wang H, Lai C, Wang L, Deng Y . Comparative proteomic analysis of human SH-SY5Y neuroblastoma cells under simulated microgravity. Astrobiology 2013; 13: 143–150.
Marks P, Arai M . Advillin (p92): a new member of the gelsolin/villin family of actin regulatory proteins. J Cell 1998; 2136: 2129–2136.
Meloni MA, Galleri G, Pani G, Saba A, Pippia P, Cogoli-Greuter M . Space flight affects motility and cytoskeletal structures in human monocyte cell line J-111. Cytoskeleton 2011; 68: 125–137.
Ulbrich C, Pietsch J, Grosse J, Schulz H, Saar K, Hübner N et al. Cellular physiology biochemistry and biochemistr y differential gene regulation under altered gravity conditions in follicular thyroid cancer cells: relationship between the extracellular matrix and the cytoskeleton. Cell Physiol Biochem 2011; 28: 185–198.
Grosse J, Wehland M, Pietsch J, Ma X, Ulbrich C, Schulz H et al. Short-term weightlessness produced by parabolic flight maneuvers altered gene expression patterns in human endothelial cells. FASEB J 2012; 26: 639–655.
Ferl RJ, Koh J, Denison F, Paul AL . Spaceflight induces specific alterations in the proteomes of Arabidopsis . Astrobiology 2015; 15: 32–56.
Fitts R, Riley D, Widrick J . Invited review: microgravity and skeletal muscle. J Appl Physiol 2000; 89: 823–839.
Röper K, Mao Y, Brown NH . Contribution of sequence variation in Drosophila actins to their incorporation into actin-based structures in vivo . J Cell Sci 2005; 118: 3937–3948.
Sörensen JG, Kristensen TN, Loeschcke V . The evolutionary and ecological role of heat shock proteins. Ecol Lett 2003; 6: 1025–1037.
Kregel KC . Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J Appl Physiol 2002; 92: 2177–2186.
Effertz C, von Elert E . Light intensity controls anti-predator defences in Daphnia: the suppression of life-history changes. Proc Biol Sci 2014; 281: 20133250.
Haap T, Triebskorn R, Köhler HR . Acute effects of diclofenac and DMSO to Daphnia magna: Immobilisation and hsp70-induction. Chemosphere 2008; 73: 353–359.
Rainville LC, Carolan D, Varela AC, Doyle H, Sheehan D . Proteomic evaluation of citrate-coated silver nanoparticles toxicity in Daphnia magna . Analyst 2014; 139: 1678–1686.
Zupanska AK, Denison FC, Ferl RJ, Paul AL . Spaceflight engages heat shock protein and other molecular chaperone genes in tissue culture cells of Arabidopsis thaliana . Am J Bot 2013; 100: 235–248.
Taylor K, Kleinhesselink K, George MD, Morgan R, Smallwood T, Hammonds AS et al. Toll mediated infection response is altered by gravity and spaceflight in Drosophila . PloS One 2014; 9: e86485.
Ingber D . How cells (might) sense microgravity. FASEB J 1999; 13: S3–S15.
Dunn aY, Melville MW, Frydman J . Review: cellular substrates of the eukaryotic chaperonin TRiC/CCT. J Struct Biol 2001; 135: 176–184.
Wilkinson B, Gilbert HF . Protein disulfide isomerase. Biochim Biophys Acta 2004; 1699: 35–44.
Wang H, Hui QZ, Sha W, Zeng R, Qi CX . A proteomic approach to analysing responses of Arabidopsis thaliana callus cells to clinostat rotation. J Exp Bot 2006; 57: 827–835.
Tan C, Wang H, Zhang Y, Qi B, Xu G, Zheng H . A proteomic approach to analyzing responses of Arabidopsis thaliana root cells to different gravitational conditions using an agravitropic mutant, pin2 and its wild type. Proteome Sci 2011; 9: 72.
Crabbé A, Schurr MJ, Monsieurs P, Morici L, Schurr J, Wilson JW et al. Transcriptional and proteomic responses of Pseudomonas aeruginosa PAO1 to spaceflight conditions involve Hfq regulation and reveal a role for oxygen. Appl Environ Microbiol 2011; 77: 1221–1230.
Wilson JW, Ott CM, Höner zu Bentrup K, Ramamurthy R, Quick L, Porwollik S et al. Space flight alters bacterial gene expression and virulence and reveals a role for global regulator Hfq. Proc Natl Acad Sci USA 2007; 104: 16299–16304.
Wilson JW, Ott CM, Quick L, Davis R, zu Bentrup KH, Crabbé A et al. Media ion composition controls regulatory and virulence response of Salmonella in spaceflight. PLoS One 2008; 3.
Lamkemeyer T, Zeis B, Paul R . Temperature acclimation influences temperature-related behaviour as well as oxygen-transport physiology and biochemistry in the water flea Daphnia magna . Can J Zool 2003; 81: 237–249.
Gerke P, Börding C, Zeis B, Paul RJ . Adaptive haemoglobin gene control in Daphnia pulex at different oxygen and temperature conditions. Comp Biochem Physiol A Physiol 2011; 159: 56–65.
Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 2014; 32: 223–226.
Acknowledgements
Our work benefits from and contributes to the Daphnia Genomics Consortium. We thank Quirin Herzog for providing D. magna photographs. We gratefully acknowledge the financial support of BMWi and DLR (Grant No. 50WB1029), as well as the ESA-GBF-program (Contract No. 4000103571).
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium57 via the PRIDE partner repository with the data set identifier PXD002096.
Author information
Authors and Affiliations
Contributions
CL, RH, and KS designed the study. BT performed clinorotation and proteomic experiments. BT and KAO analyzed proteomic data. KAO conducted further bioinformatical analysis. TF supervised mass spectrometry analysis. KAO and BT wrote the first draft of the manuscript, and CL, TF, RH, KS, and GJA contributed substantially to revisions. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Trotter, B., Otte, K., Schoppmann, K. et al. The influence of simulated microgravity on the proteome of Daphnia magna. npj Microgravity 1, 15016 (2015). https://doi.org/10.1038/npjmgrav.2015.16
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/npjmgrav.2015.16
This article is cited by
-
Proteomic analysis in the model organism Daphnia has the potential to unravel molecular pathways involved in phenotypic changes in response to changing environmental conditions
Hydrobiologia (2019)
-
Response of Haloalkaliphilic Archaeon Natronococcus Jeotgali RR17 to Hypergravity
Microgravity Science and Technology (2017)
-
Response of extreme haloarchaeon Haloarcula argentinensis RR10 to simulated microgravity in clinorotation
3 Biotech (2017)