Abstract
Down syndrome (DS) is the most common genetic cause of intellectual disability (ID) and in the majority of cases is the result of complete trisomy 21. The hypothesis that the characteristic DS clinical features are due to a single DS critical region (DSCR) at distal chromosome 21q has been refuted by recently reported segmental trisomy 21 cases characterised by microarray-based comparative genomic hybridisation (aCGH). These rare cases have implicated multiple regions on chromosome 21 in the aetiology of distinct features of DS; however, the map of chromosome 21 copy-number aberrations and their associated phenotypes remains incompletely defined. We report a child with ID who was deemed very high risk for DS on antenatal screening (1 in 13) and has partial, but distinct, dysmorphologic features of DS without congenital heart disease (CHD). Oligonucleotide aCGH testing of the proband detected a previously unreported de novo 2.78-Mb duplication on chromosome 21q22.11 that includes 16 genes; however, this aberration does not harbour any of the historical DSCR genes (APP, DSCR1, DYRK1A and DSCAM). This informative case implicates previously under-recognised candidate genes (SOD1, SYNJ1 and ITSN1) in the pathogenesis of specific DS clinical features and supports a critical region for CHD located more distal on chromosome 21q. In addition, this unique case illustrates how the increasing resolution of microarray and high-throughput sequencing technologies can continue to reveal new biology and enhance understanding of widely studied genetic diseases that were originally described over 50 years ago.
Similar content being viewed by others
Introduction
Down syndrome (DS) is one of the prototypical disorders of human aneuploidy and copy-number variation based on its characteristic clinical presentation, the identification of trisomy 21 as the hallmark cytogenetic abnormality, and the continued study of chromosome 21 candidate genes for correlation with specific DS clinical features. The hypothesis that most aspects of the DS phenotype are owing to a single DS critical region (DSCR) at distal chromosome 21q was originally proposed in the 1970s.1–3 However, the continued advances in molecular cytogenetic technologies (e.g., fluorescence in situ hybridisation (FISH) and microarray-based comparative genomic hybridisation (aCGH)) have enabled greater resolution of segmental trisomy 21 aberrations and subsequent correlation studies between candidate genes and specific DS clinical features. Notably, a DSCR thought to be responsible for key DS clinical features has been defined by a region on chromosome 21q22.12-q22.2 containing DSCR1 (RCAN1), DYRK1A, DSCAM and APP.4–6 However, recent studies of segmental trisomy 21 cases characterised by FISH and/or aCGH indicate that different features of the DS phenotype are likely attributable to several distinct genomic regions on chromosome 21 and not just a single DSCR.7,8
Despite the recent advances in ascribing DS clinical features to candidate genes and regions,7,8 the map of chromosome 21 copy-number aberrations and their associated DS clinical features remains incompletely defined. As such, the paucity of informative segmental trisomy 21 cases reported in the literature prompted our case report of a unique de novo 2.78-Mb duplication of chromosome 21q22.11 in a patient with a partial trisomy 21 phenotype. This case offers valuable insights into previously under-recognised candidate genes and regions potentially implicated in features of the DS phenotype.9
Results
Prenatal clinical history
The patient was born to a 36-year old mother, whose pregnancy had been calculated as high risk for DS (1 in 13) based on maternal age and an abnormal maternal quad screen (alfa-fetoprotein (AFP) 16.3 ng/ml, 0.47 MOM; hCG 75,636 mIU/ml, 0.74 MOM; uE3 0.61 ng/ml, 0.74 MOM; DIA 372 pg/ml; 1.72 MOM) at 15 weeks gestation; however, amniocentesis was declined. A two-vessel umbilical cord, abnormal fifth digits bilaterally, and normal cardiac anatomy were noted on antenatal ultrasound at 16 weeks gestation.
Postnatal clinical evaluation
The patient was referred to Medical Genetics at 4 years of age with physical features suggestive of DS. Height, weight and head circumference at most recent follow-up (age 5 years) were on the 3rd, 43rd and 30th centiles for age, respectively. Examination revealed a round, flat face with upslanting palpebral fissues, prominent epicanthae, flat nasal bridge (Figure 1a) and mild macroglossia. Ears had a normal appearance. Hands were broad and small with mild bilateral fifth digit clinodactyly and bilateral single transverse palmar creases, and feet showed wide interspaces between first and second toes bilaterally (Figure 1b,c). Despite the presence of mild to moderate developmental delays, the family declined a formal neuropsychologic characterisation of developmental deficits. Echocardiography showed no evidence of congenital heart disease (CHD). The patient lacked the happy, sociable affect observed in many children with DS. There was no history of intestinal atresia or other gastrointestinal malformations. Thyroid studies were normal, as was her tone, with no abnormal joint laxity or herniae.
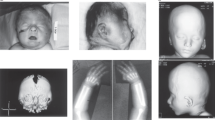
Phenotypic features of the proband and the patient reported by Huret et al.,23 and an illustration of the 21q21.2-q22.2 region highlighting the identified 2.78-Mb duplication and other reported segmental 21q duplication cases characterised by aCGH. (a–c) The proband at 5 years of age. Notable features include upslanting palpebral fissures, flat facies, prominent epicanthal folds and a flat nasal bridge. She also has bilateral single palmar creases and wide interspaces between the hallux and second digits on both feet. She has an ID, but lacks the ‘happy personality’ seen in many children with DS. Brachycephaly, protruding tongue and CHD were not present. (d) Oligonucleotide aCGH results of chromosome 21 in the proband. Coloured dots represent oligonucleotide aCGH probes plotted by their log2 ratios. The 2.78-Mb duplication at 21q22.11 is identifiable by the dense green probes with an average log2 ratio of ~0.6, indicating a single copy-number gain. (e) Chromosomal location of the 2.78-Mb duplication identified in the proband (light blue bar) compared with other reported interstitial copy-number aberrations characterised by aCGH with at least one breakpoint within 21q21.3-q22.2. Blue bands indicate duplications (partial trisomy), purple bands indicate triplications (partial tetrasomy) and red bands indicate deletions (partial monosomy). Black arrows indicate aberrations that extend proximally (left arrow) or distally (right arrow) beyond the limits of the region depicted in the figure. Genes previously identified as key critical genes in the overall DS phenotype are indicated by vertical pink translucent bars. (f) A French patient reported by Huret et al. in 1987 and photographed at 18 months of age has a similar facial appearance to our patient, lacked CHD and had a cytogenetically undetectable 21q duplication that likely involved SOD1 (see Discussion). Figure 1f reproduced from ref. 23 with permission from Springer, copyright 1987.
Clinical cytogenomic and molecular genetic analyses
Cytogenetic testing demonstrated a 46,XX normal female karyotype; however, aCGH detected a 2.78-Mb duplication of 21q22.11 (chr21:32,583,901–35,355,969; hg19) that included 16 genes (TIAM1, SOD1, HUNK, MRAP, URB1, SYNJ1, C21ORF59, OLIG2, OLIG1, IFNAR2, IFNAR1, GART, SON, DONSON, CRYZL1 and ITSN1; Figure 1d). This region has only minimal overlap with smaller copy-number variations reported among healthy individuals in the Database of Genomic Variants,10 and no duplications of comparable size or coordinates are currently reported in the ClinGen11 and DECIPHER12 databases. The 21q22.11 aberration was confirmed in the proband by interphase FISH but not detected in either parent, indicating that the 2.78-Mb duplication occurred de novo in the proband. Metaphase FISH analysis was not possible to perform so it is not currently known whether this duplication is in tandem or inserted elsewhere in the genome. Taken together, these data indicated that the de novo 2.78-Mb duplication is pathogenic.
In addition, aCGH of the proband also identified a 31.0-kb deletion of 4p16.2 (chr4:5,564,364–5,595,303; hg19) that included exons 17–22 of EVC2. This deletion was confirmed and subsequently detected in the healthy father by qPCR, suggesting that it is a paternally inherited loss-of-function allele for autosomal recessive Ellis van Creveld syndrome (EVCS). A search for physical signs of EVCS, which includes acromelic dwarfism, polydactyly, CHD, oral frenulae and dental/nail anomalies, was negative. In addition, Sanger sequencing of the EVC2 coding region in the proband did not detect any pathogenic mutations or variants of uncertain significance. Weyers acrofacial dysostosis (WAD) is characterised by dental/nail abnormalities and postaxial polydactyly, and is caused by dominant single-nucleotide mutations in the 3′ region of the last exon of EVC2,13–15 which escape nonsense-mediated messenger RNA decay and encode a dominant negative mutant EVC2 polypeptide.13,14 Furthermore, no multiexon deletions of EVC2 have been previously reported among patients with WAD, further supporting the identified 31.0-kb deletion as a recessive allele for EVCS in the proband and father.
Although stature was short, the proband was taller than that would be expected for EVCS and approximated the 80th centile in height for females with DS. Parents and the younger sister of the proband showed no facial or other physical features suggestive of DS or EVCS/WAD, and there was no family history of learning disability.
Discussion
We report the case of a child with clear neurodevelopmental and partial dysmorphologic features of DS, but without CHD, and who harbours a previously unreported de novo 2.78-Mb duplication of chromosome 21q22.11. Importantly, this aberration does not include APP, DSCR1, DYRK1A or DSCAM, which further argues against the hypothesis of a single DSCR responsible for the primary features of DS;4–8 however, the unique size and location of this duplication also supports a role for overexpression of other previously under-recognised genes in the aetiology of DS. To frame this aberration in context with other reported segmental trisomy 21 aberrations, previously reported patients with DS or partial DS phenotypes and interstitial duplications distal to 21q21.3 characterised by aCGH6–8,16–19 are summarised in Figure 1e. We excluded a recently reported case of a partial 21q duplication in an adult20 as the phenotype was likely confounded by a co-existent 2.2-Mb deletion of 7q36 in that patient.
One of the first informative cases of segmental trisomy 21 characterised by aCGH involved a maternally inherited 4.3-Mb duplication of 21q22.13-q22.2 that included DRYK1A, but not DSCR1 or DSCAM.6 The mother and elder daughter were both of short stature, had mild learning disability and had a craniofacial phenotype consistent with DS; the younger daughter, who died in the neonatal period, also had characteristic facial features. Notably, neither daughter nor the mother had CHD, prompting the conclusion that the DS facies is dependent on duplication of DYRK1A and that a more telomeric region (including DSCAM) is likely implicated in the characteristic CHD observed in ‘typical’ DS.
Additional segmental trisomy 21 cases analysed by BAC aCGH further ruled out a single DSCR being responsible for the major features of DS and narrowed many DS phenotypes to a region between 34 and 41 Mb on chromosome 21.7 This important case series also indicated that short stature and abnormal dermatoglyphics were likely due to gene regions lying outside of this interval.7 Similarly, Korbel et al.8 assessed the clinical phenotypes of 30 patients with segmental trisomy 21 and correlated clinical features with duplications defined by high-resolution oligonucleotide aCGH, which narrowed a putative CHD region to a 1.77-Mb interval telomeric to DSCR1 and DYRK1A, but including DSCAM, RIPK4 and ZBTB21. Notably, critical regions for Alzheimer’s disease (AD) and intellectual ability (ID) were also interrogated, which supported a role for overexpression of APP in AD but not in ID. Interestingly, although both DSCR1 and DYRK1A may have roles in the pathogenesis of ID, a necessary synergistic contribution of these genes to ID or CHD has not been definitively demonstrated, as some patients with ID and partial 21q duplication were trisomic for only one of either gene.6,8 The chromosomal map devised by Korbel et al.8 based on several patients with duplicated APP and normal copy number of other historically reported critical genes further suggests that more than one critical region for ID exists and argues against an essential role of APP in DS-associated ID. Moreover, a recently reported patient with an intestitial duplication involving APP and surrounding genes, but not extending distally as far as DSCR1 and observed together with a small proximal 21q triplication, had mild speech delay but no other documented developmental delays, together with macroglossia and mild foot dysmorphology.21
Our patient does not have CHD, but has ID, a facial gestalt that clearly shares features with DS, as well as single palmar creases and wide digital interspaces. Importantly, her interstitial chromosome 21q22.11 duplication is 5.3 Mb distal to APP, and 55 kb, 3.4 and >6 Mb proximal to DSCR1, DYRK1A and DSCAM, respectively (light blue bar, Figure 1e). This relatively small duplication supports previous studies that refuted exclusive roles for increased dosage of DSCR1, DYRK1A and DSCAM in the pathogenesis of neurodevelopmental and dysmorphologic aspects of the DS phenotype6–8 and reveals additional candidate genes for DS clinical features. The absence of CHD in our patient corroborates previous studies that concluded that a critical region for CHD is located distal to the identified 2.78-Mb duplication of chromosome 21q22.11, possibly including DSCAM as previously suggested.6,8
Among the OMIM-annotated genes in the identified duplication, there are few clear candidates for genes that explain the elements of our patient’s phenotype. However, previous work has suggested that overexpression of SOD1 may contribute to some aspects of DS, and it was demonstrated over 30 years ago that SOD1 activity in fetal brain is enhanced in trisomy 21.22 Soon afterwards, Huret et al.23 reported a male patient with many aspects of DS, including the characteristic facial gestalt and ID. The patient’s facial appearance (Figure 1f, reproduced with permission), was similar to that of our patient and neither had CHD. Notably, he was disomic for chromosome 21, but using superoxide dismutase enzyme assays, DNA studies and in situ hybridisation techniques, the authors concluded that he harboured a microduplication at 21q21-21q22.1 that included SOD1 but was below the level of traditional karyotype resolution.23 In Drosophila, increased SOD1 expression has been implicated in hyperphosphorylation of tau, which may serve as an important factor for AD susceptibility in children with DS.24 and abnormal SOD1 expression may also perturb oxidative stress responses, which are protective against AD.25
Another candidate gene within the identified 2.78-Mb duplication that may also contribute to the DS phenotype when overexpressed is SYNJ1, mutations in which causes an autosomal recessive Parkinson’s disease phenotype.26 This is supported by a transgenic murine model study, which suggested that appropriate Synj1 dosage is important for normal brain development and that overexpression of Synj1 is implicated in the brain dysfunction observed in DS.27 Pucharcos et al.28 previously demonstrated in a mouse model that itsn1 is expressed in both proliferating and differentiating neurons, and proposed that ITSN1 overexpression may contribute to DS pathogenesis.
In conclusion, the identified 2.78-Mb duplication at 21q22.11 supports roles for SOD1, SYNJ1 and/or ITSN1 overexpression in the partial trisomy 21 phenotype, specifically correlating with characteristic DS facial features, hand and foot dysmorphology, and ID. This unique clinical case again underscores the utility of cytogenomic approaches to the complex DS phenotype as well as other Mendelian disorders, and indicates that the ongoing advances in genomic technologies, including whole-genome sequencing, will continue to clarify unanswered questions in clinical genetics as well as illuminate new hypotheses and uncertainties in the evolving discipline of genomic medicine. Each newly identified partial 21q duplication patient should be reported in order to advance our understanding of genotype–phenotype correlations in the DS phenotype.
Materials and methods
Clinical evaluation
Clinical evaluation was performed at the Medical Genetics clinic at Children’s Hospital Colorado by a board-certified clinical geneticist and paediatrician (J.D.W-A.) and genetic counselor (A.K.T.). Informed consent was obtained from the mother of the patient for their participation in this study, including the use of medical information and the publication of the patient's photograph (Figures 1a–c). Methods were performed in accordance with relevant regulations and guidelines.
Clinical cytogenetic and molecular genetic testing
Clinical cytogenetic and cytogenomic testing included karyotype analysis performed at the Colorado Genetics Laboratory (CGL) at the University of Colorado, and aCGH testing using the 400 K CMA-COMP array v9.1 (Agilent Technologies, Inc., Santa Clara, CA, USA) performed at the Baylor Miraca Genetics Laboratories. The identified chromosome 21q22.11 duplication was further interrogated at the Baylor Miraca Genetics Laboratories by interphase FISH using the RP11-484I12 BAC probe. The identified chromosome 4p16.2 deletion was interrogated at the DNA Diagnostic Laboratory at the University of Colorado Denver using a dual Taqman probe quantitative PCR (qPCR) assay (Roche Diagnostics Corporation, Indianapolis, IN, USA) with 4p16.2 region-specific primers (forward: 5′- GTGCACTCACATTGCACCAT-3′; reverse: 5′- CATGACTCTGTCTTGCCTGGT-3′) and a Universal Probe (5′- CATCCAGC-3′). In addition, EVC2 Sanger sequencing was performed at the DNA Diagnostic Laboratory at the University of Colorado, Denver, using standard protocols.
Change history
25 August 2017
A correction has been published for this paper. The error has been fixed in both the HTML and PDF versions of the paper.
References
Niebuhr, E. Down's syndrome. The possibility of a pathogenetic segment on chromosome no. 21. Humangenetik 21, 99–101 (1974).
Sinet, P. M. et al. Molecular mapping of the Down syndrome phenotype on chromosome 21. Prog. Clin. Biol. Res. 384, 63–86 (1993).
Hagemeijer, A. & Smit, E. M. Partial trisomy 21. Further evidence that trisomy of band 21q22 is essential for Down's phenotype. Hum. Genet. 38, 15–23 (1977).
Delabar, J. M. et al. Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur. J. Hum. Genet. 1, 114–124 (1993).
Arron, J. R. et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature 441, 595–600 (2006).
Ronan, A. et al. Familial 4.3 Mb duplication of 21q22 sheds new light on the Down syndrome critical region. J. Med. Genet. 44, 448–451 (2007).
Lyle, R. et al. Genotype-phenotype correlations in Down syndrome identified by array CGH in 30 cases of partial trisomy and partial monosomy chromosome 21. Eur. J. Hum. Genet. 17, 454–466 (2009).
Korbel, J. O. et al. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc. Natl Acad. Sci. USA 106, 12031–12036 (2009).
Lejeune, J., Gautier, M. & Turpin, R. [Study of somatic chromosomes from 9 mongoloid children]. C. R. Hebd. Seances Acad. Sci. 248, 1721–1722 (1959).
MacDonald, J. R., Ziman, R., Yuen, R. K., Feuk, L. & Scherer, S. W. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 42, D986–D992 (2014).
Rehm, H. L. et al. ClinGen—the clinical genome resource. N. Engl. J. Med. 372, 2235–2242 (2015).
Firth, H. V. et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 84, 524–533 (2009).
Valencia, M. et al. Widening the mutation spectrum of EVC and EVC2: ectopic expression of Weyer variants in NIH 3T3 fibroblasts disrupts Hedgehog signaling. Hum. Mutat. 30, 1667–1675 (2009).
D'Asdia, M. C. et al. Novel and recurrent EVC and EVC2 mutations in Ellis-van Creveld syndrome and Weyers acrofacial dyostosis. Eur. J. Med. Genet. 56, 80–87 (2013).
Ye, X. et al. A novel heterozygous deletion in the EVC2 gene causes Weyers acrofacial dysostosis. Hum. Genet. 119, 199–205 (2006).
Chen, C. P. et al. A de novo duplication of chromosome 21q22.11-->qter associated with Down syndrome: prenatal diagnosis, molecular cytogenetic characterization and fetal ultrasound findings. Taiwan J. Obstet. Gynecol. 50, 492–498 (2011).
Eggermann, T. et al. Identification of a 21q22 duplication in a Silver-Russell syndrome patient further narrows down the Down syndrome critical region. Am. J. Med. Genet. A 152A, 356–359 (2010).
Qi, Q. et al. A rare de novo duplication of chromosome 21q22.12 --> q22.3 with other concomitant deletion and duplication of small fragments in 21q associated with Down syndrome: Prenatal diagnosis, molecular cytogenetic characterization. Mol. Cytogenet. 6, 11 (2013).
Vaglio, A. et al. Clinical, cytogenetic, and molecular characterization of a girl with some clinical features of Down syndrome resulting from a pure partial trisomy 21q22.11-qter due to a de novo intrachromosomal duplication. Genet. Test Mol. Biomarkers 14, 57–65 (2010).
Papoulidis, I. et al. A patient with partial trisomy 21 and 7q deletion expresses mild Down syndrome phenotype. Gene 536, 441–443 (2014).
Capkova, P., Misovicova, N. & Vrbicka, D. Partial trisomy and tetrasomy of chromosome 21 without Down Syndrome phenotype and short overview of genotype-phenotype correlation. A case report. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc. Czech Repub. 158, 321–325 (2014).
Brooksbank, B. W. & Balazs, R. Superoxide dismutase and lipoperoxidation in Down's syndrome fetal brain. Lancet 1, 881–882 (1983).
Huret, J. L. et al. Down syndrome with duplication of a region of chromosome 21 containing the CuZn superoxide dismutase gene without detectable karyotypic abnormality. Hum. Genet. 75, 251–257 (1987).
Huang, Y., Wu, Z. & Zhou, B. hSOD1 promotes tau phosphorylation and toxicity in the Drosophila model. J. Alzheimers Dis. 45, 235–244 (2015).
Schupf, N. et al. Candidate genes for Alzheimer's disease are associated with individual differences in plasma levels of beta amyloid peptides in adults with Down syndrome. Neurobiol. Aging 36, 2907, e1-10 (2015).
Quadri, M. et al. Mutation in the SYNJ1 gene associated with autosomal recessive, early-onset Parkinsonism. Hum. Mutat. 34, 1208–1215 (2013).
Voronov, S. V. et al. Synaptojanin 1-linked phosphoinositide dyshomeostasis and cognitive deficits in mouse models of Down's syndrome. Proc. Natl Acad. Sci. USA 105, 9415–9420 (2008).
Pucharcos, C. et al. Alu-splice cloning of human Intersectin (ITSN), a putative multivalent binding protein expressed in proliferating and differentiating neurons and overexpressed in Down syndrome. Eur. J. Hum. Genet. 7, 704–712 (1999).
Acknowledgements
We thank the proband and family for supporting publication of this case report. KNM. gratefully acknowledges financial support from the Linda Crnic Institute for Down Syndrome. SAS. was supported, in part, by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH) through grant K23 GM104401.
Author information
Authors and Affiliations
Contributions
JDW-A. oversaw the clinical workup, conceived the case report, reviewed the literature, analysed the data, and drafted and revised the manuscript; AKT. executed the clinical workup, analysed the data and revised the manuscript; KNM. reviewed the literature, analysed the data and revised the manuscript; NLM. reviewed the literature, analysed the data and revised the manuscript; and SAS. assisted in conceiving the case report, reviewed the literature, analysed the data, and drafted and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Weisfeld-Adams, J., Tkachuk, A., Maclean, K. et al. A de novo 2.78-Mb duplication on chromosome 21q22.11 implicates candidate genes in the partial trisomy 21 phenotype. npj Genomic Med 1, 16003 (2016). https://doi.org/10.1038/npjgenmed.2016.3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/npjgenmed.2016.3
This article is cited by
-
Down Syndrome: how to communicate the diagnosis
Italian Journal of Pediatrics (2023)
-
Investigation of copy number variations on chromosome 21 detected by comparative genomic hybridization (CGH) microarray in patients with congenital anomalies
Molecular Cytogenetics (2018)