Abstract
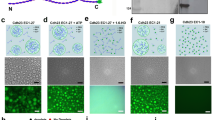
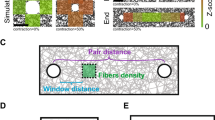
The integrity of living tissues is maintained by adhesion domains of trans-bonds formed between cadherin proteins residing on opposing membranes of neighbouring cells. These domains are stabilized by lateral cis-interactions between the cadherins on the same cell. However, the origin of cis-interactions remains perplexing since they are detected only in the context of trans-bonds. By combining experimental, analytical and computational approaches, we identify bending fluctuations of membranes as a source of long-range cis-interactions, and a regulator of trans-interactions. Specifically, nanometric membrane bending and fluctuations introduce cooperative effects that modulate the affinity and binding/unbinding rates for trans-dimerization, dramatically affecting the nucleation and growth of adhesion domains. Importantly, this regulation relies on physical principles and not on details of protein–protein interactions. These omnipresent fluctuations can thus act as a generic control mechanism in all types of cell adhesion, suggesting a hitherto unknown physiological role for recently identified active fluctuations of cellular membranes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Leckband, D. & Sivasankar, S. Cadherin recognition and adhesion. Curr. Opin. Cell Biol. 24, 620–627 (2012).
Lecuit, T. & Yap, A. S. E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat. Cell Biol. 17, 533–539 (2015).
Wu, Y., Vendome, J., Shapiro, L., Ben-Shaul, A. & Honig, B. Transforming binding affinities from three dimensions to two with application to cadherin clustering. Nature 475, 510–513 (2011).
Volk, T., Cohen, O. & Geiger, B. Formation of heterotypic adherens-type junctions between L-CAM-containing liver cells and A-CAM-containing lens cells. Cell 50, 987–994 (1987).
Ayalon, O. et al. Spatial and temporal relationships between cadherins and pecam-1 in cell–cell junctions of human endothelial cells. J. Cell Biol. 126, 247–258 (1994).
Strale, P.-O. et al. The formation of ordered nanoclusters controls cadherin anchoring to actin and cell–cell contact fluidity. J. Cell Biol. 210, 333–346 (2015).
Engl, W. et al. Actin dynamics modulate mechanosensitive immobilization of E-cadherin at adherens junctions. Nat. Cell Biol. 16, 584–591 (2014).
Bello, S. M. et al. Catenin-dependent cadherin function drives divisional segregation of spinal motor neurons. J. Neurosci. 32, 490–505 (2012).
Katsamba, P. et al. Linking molecular affinity and cellular specificity in cadherin-mediated adhesion. Proc. Natl Acad. Sci. USA 106, 11594–11599 (2009).
Gumbiner, B. M. Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 6, 622–634 (2005).
Jeanes, A., Gottardi, C. J. & Yap, A. S. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene 27, 6920–6929 (2008).
Biswas, K. H. et al. E-cadherin junction formation involves an active kinetic nucleation process. Proc. Natl Acad. Sci. USA 112, 10932–10937 (2015).
Rakshit, S. & Sivasankar, S. Biomechanics of cell adhesion: how force regulates the lifetime of adhesive bonds at the single molecule level. Phys. Chem. Chem. Phys. 16, 2211–2223 (2014).
Sivasankar, S. et al. Characterizing the initial encounter complex in cadherin adhesion. Structure 17, 1075–1081 (2009).
Harrison, O. J. et al. Two-step adhesive binding by classical cadherins. Nat. Struct. Mol. Biol. 17, 348–357 (2010).
Boggon, T. J. et al. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science 296, 1308–1313 (2002).
Fenz, S. F. & Sengupta, K. Giant vesicles as cell models. Integr. Biol. 4, 982–995 (2012).
Vendome, J. et al. Structural and energetic determinants of adhesive binding specificity in type I cadherins. Proc. Natl Acad. Sci. USA 111, E4175–E4184 (2014).
Harrison, O. J. et al. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure 19, 244–256 (2011).
Taveau, J.-C. et al. Structure of artificial and natural VE-cadherin-based adherens junctions. Biochem. Soc. Trans. 36, 189–193 (2008).
Hong, S., Troyanovsky, R. B. & Troyanovsky, S. M. Spontaneous assembly and active disassembly balance adherens junction homeostasis. Proc. Natl Acad. Sci. USA 107, 3528–3533 (2010).
Ozaki, C. et al. The extracellular domains of E- and N-cadherin determine the scattered punctate localization in epithelial cells and the cytoplasmic domains modulate the localization. J. Biochem. 147, 415–425 (2010).
Hong, S., Troyanovsky, R. B. & Troyanovsky, S. M. Binding to F-actin guides cadherin cluster assembly, stability, and movement. J. Cell Biol. 201, 131–143 (2013).
Wu, Y. et al. Cooperativity between trans and cis interactions in cadherin-mediated junction formation. Proc. Natl Acad. Sci. USA 107, 17592–17597 (2010).
Brasch, J. et al. Thinking outside the cell: how cadherins drive adhesion. Trends Cell Biol. 22, 299–310 (2012).
Fenz, S. F., Merkel, R. & Sengupta, K. Diffusion and intermembrane distance: case study of avidin and E-cadherin mediated adhesion. Langmuir 25, 1074–1085 (2009).
Limozin, L. & Sengupta, K. Quantitative reflection interference contrast microscopy (RICM) in soft matter and cell adhesion. ChemPhysChem 10, 2752–2768 (2009).
Fenz, S. F. et al. Inter-membrane adhesion mediated by mobile linkers: effect of receptor shortage. Soft Matter 7, 952–962 (2011).
Sengupta, K. & Limozin, L. Adhesion of soft membranes controlled by tension and interfacial polymers. Phys. Rev. Lett. 104, 088101 (2010).
Schmidt, D. et al. Signature of a nonharmonic potential as revealed from a consistent shape and fluctuation analysis of an adherent membrane. Phys. Rev. X 4, 021023 (2014).
Rädler, J. O. et al. Fluctuation analysis of tension-controlled undulation forces between giant vesicles and solid substrates. Phys. Rev. E 51, 4526–4536 (1995).
Seifert, U. Configuration of fluid membranes and vesicles. Adv. Phys. 46, 13–137 (1997).
Schmidt, D. et al. Coexistence of dilute and densely packed domains of ligand-receptor bonds in membrane adhesion. Europhys. Lett. 99, 38003 (2012).
Bazellières, E. et al. Control of cell–cell forces and collective cell dynamics by the intercellular adhesome. Nat. Cell Biol. 17, 409–420 (2015).
Bell, G. I. Models for the specific adhesion of cells to cells. Science 200, 618–627 (1978).
Dembo, M. et al. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc. R. Soc. Lond. B 234, 55–83 (1988).
Bihr, T., Seifert, U. & Smith, A.-S. Multiscale approaches to protein-mediated interactions between membranes—relating microscopic and macroscopic dynamics in radially growing adhesions. New J. Phys. 17, 083016 (2015).
Monzel, C. et al. Measuring fast stochastic displacements of bio-membranes with dynamic optical displacement spectroscopy. Nat. Commun. 6, 1221–1229 (2015).
Turlier, H. et al. Equilibrium physics breakdown reveals the active nature of red blood cell flickering. Nat. Phys. 12, 513–516 (2016).
Bihr, T., Seifert, U. & Smith, A.-S. Nucleation of ligand-receptor domains in membrane adhesion. Phys. Rev. Lett. 109, 258101 (2012).
Fenz, S. F. et al. Switching from ultraweak to strong adhesion. Adv. Mater. 23, 2622–2626 (2011).
Adams, C. L. et al. Mechanisms of epithelial cell–cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin-green fluorescent protein. J. Cell Biol. 142, 1105–1119 (1998).
Perez, T.-D. et al. Immediate-early signaling induced by E-cadherin engagement and adhesion. J. Biol. Chem. 283, 5014–5022 (2008).
Smith, A.-S. et al. Force-induced growth of adhesion domains is controlled by receptor mobility. Proc. Natl Acad. Sci. USA 105, 6906–6911 (2008).
Schmidt, D. et al. Crowding of receptors induces ring-like adhesions in model membranes. Biochim. Biophys. Acta 1853, 2984–2991 (2015).
Dustin, M. L. et al. Identification of self through two-dimensional chemistry and synapses. Annu. Rev. Cell Dev. Biol. 17, 133–157 (2001).
Zhu, D.-M. et al. Analysis of two-dimensional dissociation constant of laterally mobile cell adhesion molecules. Biophys. J. 92, 1022–1034 (2007).
Hu, J., Lipowsky, R. & Weikl, T. R. Binding constants of membrane-anchored receptors and ligands depend strongly on the nanoscale roughness of membranes. Proc. Natl Acad. Sci. USA 110, 15283–15288 (2013).
Boulbitch, A., Guttenberg, Z. & Sackmann, E. Kinetics of membrane adhesion mediated by ligand-receptor interaction studied with a biomimetic system. Biophys. J. 81, 2743–2751 (2001).
Lorz, B. G. et al. Adhesion of giant vesicles mediated by weak binding of sialyl-lewisX to E-selectin in the presence of repelling poly(ethylene glycol) molecules. Langmuir 23, 12293–12300 (2007).
Wu, Y., Honig, B. & Ben-Shaul, A. Theory and simulations of adhesion receptor dimerization on membrane surfaces. Biophys. J. 104, 1221–1229 (2013).
Zidovska, A. & Sackmann, E. Brownian motion of nucleated cell envelopes impedes adhesion. Phys. Rev. Lett. 96, 048103 (2006).
Pierres, A. et al. How cells tiptoe on adhesive surfaces before sticking. Biophys. J. 94, 4114–4122 (2008).
Monzel, C. et al. Probing biomembrane dynamics by dual-wavelength reflection interference contrast microscopy. ChemPhysChem 10, 2828–2838 (2009).
Acknowledgements
A.-S.S. and T.B. were funded from the grant ERC StG 2013-337283 and K.S. from ERC-StG 307104FP of the European Research Council. A.-S.S. and D.S. were supported by the Research Training Group 1962 at the Friedrich-Alexander-Universität Erlangen-Nürnberg. This work has partly been supported by A∗MIDEX (no. ANR-11-IDEX-0001-02), the Croatian Science Foundation (IP-11-2013-8238 CompSoLS-MolFlex) and the BigThera project at FAU.
Author information
Authors and Affiliations
Contributions
The project was conceived and supervised by A.-S.S., K.S., U.S. and R.M. The experimental set-up was established by S.F.F. and K.S. and applied to the current problem by S.F.F. Data analysis was performed by S.F.F., D.S., K.S. and A.-S.S. The simulation set-up was built by T.B. and A.-S.S., and executed by T.B. The theoretical model was developed by D.S. and A.-S.S. with the help of T.B. and U.S. All authors contributed to the interpretation of results. The article was written by T.B., D.S., S.F.F., K.S. and A.-S.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 697 kb)
Supplementary movie
Supplementary movie 1 (MOV 3086 kb)
Supplementary movie
Supplementary movie 2 (MOV 2376 kb)
Supplementary movie
Supplementary movie 3 (MOV 4380 kb)
Supplementary movie
Supplementary movie 4 (MOV 2552 kb)
Rights and permissions
About this article
Cite this article
Fenz, S., Bihr, T., Schmidt, D. et al. Membrane fluctuations mediate lateral interaction between cadherin bonds. Nature Phys 13, 906–913 (2017). https://doi.org/10.1038/nphys4138
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nphys4138
This article is cited by
-
Measuring sub-nanometer undulations at microsecond temporal resolution with metal- and graphene-induced energy transfer spectroscopy
Nature Communications (2024)
-
Patterning and dynamics of membrane adhesion under hydraulic stress
Nature Communications (2023)
-
Thermal fluctuations of the lipid membrane determine particle uptake into Giant Unilamellar Vesicles
Nature Communications (2023)
-
Nucleation of cadherin clusters on cell-cell interfaces
Scientific Reports (2022)
-
Cell–cell interfaces as specialized compartments directing cell function
Nature Reviews Molecular Cell Biology (2020)