Abstract
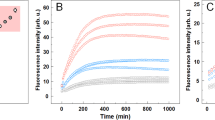
The ability of biological molecules to replicate themselves is the foundation of life, requiring a complex cellular machinery. However, a range of aberrant processes involve the self-replication of pathological protein structures without any additional assistance. One example is the autocatalytic generation of pathological protein aggregates, including amyloid fibrils, involved in neurodegenerative disorders. Here, we use computer simulations to identify the necessary requirements for the self-replication of fibrillar assemblies of proteins. We establish that a key physical determinant for this process is the affinity of proteins for the surfaces of fibrils. We find that self-replication can take place only in a very narrow regime of inter-protein interactions, implying a high level of sensitivity to system parameters and experimental conditions. We then compare our theoretical predictions with kinetic and biosensor measurements of fibrils formed from the Aβ peptide associated with Alzheimer’s disease. Our results show a quantitative connection between the kinetics of self-replication and the surface coverage of fibrils by monomeric proteins. These findings reveal the fundamental physical requirements for the formation of supra-molecular structures able to replicate themselves, and shed light on mechanisms in play in the proliferation of protein aggregates in nature.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Prusiner, S. B. Prions. Proc. Natl Acad. Sci. USA 95, 13363–13383 (1998).
Hall, D. & Edskes, H. Silent prions lying in wait: a two-hit model of prion/amyloid formation and infection. J. Mol. Biol. 336, 775–786 (2004).
Collins, S. R., Douglass, A., Vale, R. D. & Weissman, J. S. Mechanism of prion propagation: amyloid growth occurs by monomer addition. PLoS Biol. 2, e321 (2004).
Ferrone, F. A., Hofrichter, J. & Eaton, W. A. Kinetics of sickle hemoglobin polymerization: II. A double nucleation mechanism. J. Mol. Biol. 183, 611–631 (1985).
Eaton, W. A. & Hofrichter, J. Sickle cell hemoglobin polymerization. Adv. Protein Chem. 40, 263–279 (1990).
Dobson, C. M. Protein misfolding, evolution and disease. Trends Biochem. Sci. 24, 329–332 (1999).
Chiti, F. & Dobson, C. M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006).
Knowles, T. P., Vendruscolo, M. & Dobson, C. M. The amyloid state and its association with protein misfolding diseases. Nature Rev. Mol. Cell Biol. 15, 384–396 (2014).
Hortschansky, P., Schroeckh, V., Christopeit, T., Zandomeneghi, G. & Fändrich, M. The aggregation kinetics of Alzheimer’s β-amyloid peptide is controlled by stochastic nucleation. Protein Sci. 14, 1753–1759 (2005).
Cohen, S. I. et al. Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl Acad. Sci. USA 110, 9758–9763 (2013).
Padrick, S. B. & Miranker, A. D. Islet amyloid: phase partitioning and secondary nucleation are central to the mechanism of fibrillogenesis. Biochemistry 41, 4694–4703 (2002).
Ruschak, A. M. & Miranker, A. D. Fiber-dependent amyloid formation as catalysis of an existing reaction pathway. Proc. Natl Acad. Sci. USA 104, 12341–12346 (2007).
Schlamadinger, D. E. & Miranker, A. D. Fiber-dependent and -independent toxicity of islet amyloid polypeptide. Biophys. J. 107, 2559–2566 (2014).
Buell, A. K. et al. Solution conditions determine the relative importance of nucleation and growth processes in α-synuclein aggregation. Proc. Natl Acad. Sci. USA 111, 7671–7676 (2014).
Galvagnion, C. et al. Lipid vesicles trigger α-synuclein aggregation by stimulating primary nucleation. Nature Chem. Biol. 11, 229–234 (2015).
Foderà, V., Librizzi, F., Groenning, M., Van De Weert, M. & Leone, M. Secondary nucleation and accessible surface in insulin amyloid fibril formation. J. Phys. Chem. B 112, 3853–3858 (2008).
Meisl, G., Yang, X., Frohm, B., Knowles, T. P. & Linse, S. Quantitative analysis of intrinsic and extrinsic factors in the aggregation mechanism of Alzheimer-associated Aβ-peptide. Sci. Rep. 6, 18728 (2016).
Meisl, G., Yang, X., Dobson, C. M., Linse, S. & Knowles, T. P. J. A general reaction network unifies the aggregation behaviour of the Aβ42 peptide and its variants. Preprint at http://arXiv.org/abs/1604.00828 (2016).
Bieler, N. S., Knowles, T. P., Frenkel, D. & Vácha, R. Connecting macroscopic observables and microscopic assembly events in amyloid formation using coarse grained simulations. PLoS Comput. Biol. 8, e1002692 (2012).
Šarić, A., Chebaro, Y. C., Knowles, T. P. & Frenkel, D. Crucial role of nonspecific interactions in amyloid nucleation. Proc. Natl Acad. Sci. USA 111, 17869–17874 (2014).
Cohen, S. I. et al. A molecular chaperone breaks the catalytic cycle that generates toxic Aβ oligomers. Nature Struct. Mol. Biol. 22, 207–213 (2015).
Nelson, R. et al. Structure of the cross-β spine of amyloid-like fibrils. Nature 435, 773–778 (2005).
Serio, T. R. et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289, 1317–1321 (2000).
Anwar, J., Khan, S. & Lindfors, L. Secondary crystal nucleation: nuclei breeding factory uncovered. Angew. Chem. 127, 14894–14897 (2015).
Meisl, G. et al. Differences in nucleation behavior underlie the contrasting aggregation kinetics of the Aβ40 and Aβ42 peptides. Proc. Natl Acad. Sci. USA 111, 9384–9389 (2014).
Bucciantini, M. et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416, 507–511 (2002).
Haass, C. & Selkoe, D. J. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid β-peptide. Nature Rev. Mol. Cell Biol. 8, 101–112 (2007).
Walsh, D. M. et al. Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature 416, 535–539 (2002).
Zeravcic, Z. & Brenner, M. P. Self-replicating colloidal clusters. Proc. Natl Acad. Sci. USA 111, 1748–1753 (2014).
Wang, T. et al. Self-replication of information-bearing nanoscale patterns. Nature 478, 225–228 (2011).
Zhang, J. & Muthukumar, M. Simulations of nucleation and elongation of amyloid fibrils. J. Chem. Phys. 130, 035102 (2009).
Ilie, I. M., den Otter, W. K. & Briels, W. J. A coarse grained protein model with internal degrees of freedom. Application to α-synuclein aggregation. J. Chem. Phys. 144, 085103 (2016).
Ruff, K. M., Khan, S. J. & Pappu, R. V. A coarse-grained model for polyglutamine aggregation modulated by amphipathic flanking sequences. Biophys. J. 107, 1226–1235 (2014).
Ruff, K. M., Harmon, T. S. & Pappu, R. V. Camelot: a machine learning approach for coarse-grained simulations of aggregation of block-copolymeric protein sequences. J. Chem. Phys. 143, 243123 (2015).
Fitzpatrick, A. W. et al. Atomic structure and hierarchical assembly of a cross-β amyloid fibril. Proc. Natl Acad. Sci. USA 110, 5468–5473 (2013).
Serpell, L. C. Alzheimer’s amyloid fibrils: structure and assembly. Biochim. Biophys. Acta 1502, 16–30 (2000).
Davis, C. H. & Berkowitz, M. L. A molecular dynamics study of the early stages of amyloid-β (1–42) oligomerization: the role of lipid membranes. Proteins 78, 2533–2545 (2010).
Buell, A. K. et al. Detailed analysis of the energy barriers for amyloid fibril growth. Angew. Chem. Int. Ed. 51, 5247–5251 (2012).
Vácha, R. & Frenkel, D. Relation between molecular shape and the morphology of self-assembling aggregates: a simulation study. Biophys. J. 101, 1432–1439 (2011).
Fandrich, M., Fletcher, M. A. & Dobson, C. M. Amyloid fibrils from muscle myoglobin—even an ordinary globular protein can assume a rogue guise if conditions are right. Nature 410, 165–166 (2001).
Allison, J. R., Varnai, P., Dobson, C. M. & Vendruscolo, M. Determination of the free energy landscape of α-synuclein using spin label nuclear magnetic resonance measurements. J. Am. Chem. Soc. 131, 18314–18326 (2009).
Frenkel, D. & Smit, B. Understanding Molecular Simulation: From Algorithms to Applications Vol. 1 (Academic, 2001).
Michaels, T. C., Lazell, H. W., Arosio, P. & Knowles, T. P. Dynamics of protein aggregation and oligomer formation governed by secondary nucleation. J. Chem. Phys. 143, 054901 (2015).
Hellstrand, E., Boland, B., Walsh, D. M. & Linse, S. Amyloid β-protein aggregation produces highly reproducible kinetic data and occurs by a two-phase process. ACS Chem. Neurosci. 1, 13–18 (2009).
Acknowledgements
We acknowledge support from the Human Frontier Science Program and Emmanuel College (A.Š.), the Leverhulme Trust and Magdalene College (A.K.B.), St John’s College (T.C.T.M.), the Biotechnology and Biological Sciences Research Council (T.P.J.K. and C.M.D.), the Frances and Augustus Newman Foundation (T.P.J.K.), the European Research Council (T.P.J.K., T.C.T.M., S.L. and D.F.), and the Engineering and Physical Sciences Research Council (D.F.).
Author information
Authors and Affiliations
Contributions
A.Š., T.P.J.K. and D.F. conceived the project; A.Š. designed and performed the computer simulations; A.K.B. performed the SPR measurements; G.M. performed the kinetic analysis; T.C.T.M. and S.L. contributed materials and/or analysis tools, and all authors participated in interpreting the results and writing the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 4507 kb)
Rights and permissions
About this article
Cite this article
Šarić, A., Buell, A., Meisl, G. et al. Physical determinants of the self-replication of protein fibrils. Nature Phys 12, 874–880 (2016). https://doi.org/10.1038/nphys3828
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nphys3828
This article is cited by
-
Amyloid formation as a protein phase transition
Nature Reviews Physics (2023)
-
Mechanisms of enhanced aggregation and fibril formation of Parkinson’s disease-related variants of α-synuclein
Scientific Reports (2022)
-
Dynamics of oligomer populations formed during the aggregation of Alzheimer’s Aβ42 peptide
Nature Chemistry (2020)
-
Repeat domain-associated O-glycans govern PMEL fibrillar sheet architecture
Scientific Reports (2019)
-
Amyloidophilic Molecule Interactions on the Surface of Insulin Fibrils: Cooperative Binding and Fluorescence Quenching
Scientific Reports (2019)