Abstract
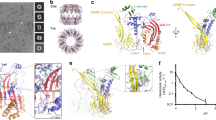
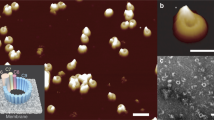
Perforin is a key protein of the vertebrate immune system. Secreted by cytotoxic lymphocytes as soluble monomers, perforin can self-assemble into oligomeric pores of 10–20 nm inner diameter in the membranes of virus-infected and cancerous cells. These large pores facilitate the entry of pro-apoptotic granzymes, thereby rapidly killing the target cell. To elucidate the pathways of perforin pore assembly, we carried out real-time atomic force microscopy and electron microscopy studies. Our experiments reveal that the pore assembly proceeds via a membrane-bound prepore intermediate state, typically consisting of up to approximately eight loosely but irreversibly assembled monomeric subunits. These short oligomers convert to more closely packed membrane nanopore assemblies, which can subsequently recruit additional prepore oligomers to grow the pore size.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Voskoboinik, I., Whisstock, J. C. & Trapani, J. A. Perforin and granzymes: function, dysfunction and human pathology. Nat. Rev. Immunol. 15, 388–400 (2016).
Reboul, C. F., Whisstock, J. C. & Dunstone, M. A. Giant MACPF/CDC pore forming toxins: a class of their own. Biochim. Biophys. Acta 1858, 475–486 (2016).
Lukoyanova, N., Hoogenboom, B. W. & Saibil, H. R. The membrane attack complex/perforin and cholesterol-dependent cytolysin (MACPF/CDC) superfamily of pore forming proteins. J. Cell Sci. 129, 2125–2133 (2016).
Shatursky, O. et al. The mechanism of membrane insertion for a cholesterol-dependent cytolysin: a novel paradigm for pore-forming toxins. Cell 99, 293–299 (1999).
Shepard, L. A., Shatursky, O., Johnson, A. E. & Tweten, R. K. The mechanism of pore assembly for a cholesterol-dependent cytolysin: formation of a large prepore complex precedes the insertion of the transmembrane β-hairpins. Biochemistry 39, 10284–10293 (2000).
Hotze, E. M. et al. Arresting pore formation of a cholesterol-dependent cytolysin by disulfide trapping synchronizes the insertion of the transmembrane β-sheet from a prepore intermediate. J Biol. Chem. 276, 8261–8268 (2001).
Czajkowsky, D. M., Hotze, E. M., Shao, Z. F. & Tweten, R. K. Vertical collapse of a cytolysin prepore moves its transmembrane β-hairpins to the membrane. EMBO J. 23, 3206–3215 (2004).
Ramachandran, R., Tweten, R. K. & Johnson, A. E. The domains of a cholesterol-dependent cytolysin undergo a major FRET-detected rearrangement during pore formation. Proc. Natl Acad. Sci. USA 102, 7139–7144 (2005).
Tilley, S. J., Orlova, E. V., Gilbert, R. J. C., Andrew, P. W. & Saibil, H. R. Structural basis of pore formation by the bacterial toxin pneumolysin. Cell 121, 247–256 (2005).
Leung, C. et al. Stepwise visualization of membrane pore formation by suilysin, a bacterial cholesterol-dependent cytolysin. eLife 3, e04247 (2014).
Sonnen, A. F. P., Plitzko, J. M. & Gilbert, R. J. C. Incomplete pneumolysin oligomers form membrane pores. Open Biol. 4, 140044 (2014).
Podobnik, M. et al. Plasticity of listeriolysin O pores and its regulation by pH and unique histidine. Sci. Rep. 5, 9623 (2015).
Mulvihill, E., van Pee, K., Mari, S. A., Muller, D. J. & Yildiz, O. Directly observing the lipid-dependent self-assembly and pore-forming mechanism of the cytolytic toxin listeriolysin O. Nano Lett. 15, 6965–6973 (2015).
Ruan, Y., Rezelj, S., Zavec, A. B., Anderluh, G. & Scheuring, S. Listeriolysin O membrane damaging activity involves arc formation and lineaction—implication for Listeria monocytogenes escape from phagocytic vacuole. PLoS Pathog. 12, e1005597 (2016).
Law, R. H. P. et al. The structural basis for membrane binding and pore formation by lymphocyte perforin. Nature 468, 447–451 (2010).
Dudkina, N. V. et al. Structure of the poly-C9 component of the complement membrane attack complex. Nat. Commun. 7, 10588 (2016).
Serna, M., Giles, J. L., Morgan, P. & Bubeck, D. Structural basis of complement membrane attack complex formation. Nat. Commun. 7, 10587 (2016).
Lukoyanova, N. et al. Conformational changes during pore formation by the perforin-related protein pleurotolysin. PLoS Biol. 13, e1002049 (2015).
Scibek, J. J., Plumb, E. P. & Sodetz, J. M. Binding of human complement C8 to C9: role of the N-terminal modules in the C8α subunit. Biochemistry 41, 14546–14551 (2002).
Aleshin, A. E. et al. Structure of complement C6 suggests a mechanism for initiation and unidirectional, sequential assembly of membrane attack complex (MAC). J. Biol. Chem. 287, 10210–10222 (2012).
Metkar, S. S. et al. Perforin oligomers form arcs in cellular membranes: a locus for intracellular delivery of granzymes. Cell Death Differ. 22, 74–85 (2015).
Praper, T. et al. Human perforin employs different avenues to damage membranes. J. Biol. Chem. 286, 2946–2955 (2011).
Lopez, J. A. et al. Perforin forms transient pores on the target cell plasma membrane to facilitate rapid access of granzymes during killer cell attack. Blood 121, 2659–2668 (2013).
Hodel, A. W., Leung, C., Dudkina, N. V., Saibil, H. R. & Hoogenboom, B. W. Atomic force microscopy of membrane pore formation by cholesterol dependent cytolysins. Curr. Opin. Struct. Biol. 39, 8–15 (2016).
Gilbert, R. J. C., Mikelj, M., Dalla Serra, M., Froelich, C. J. & Anderluh, G. Effects of MACPF/CDC proteins on lipid membranes. Cell. Mol. Life Sci. 70, 2083–2098 (2013).
Voskoboinik, I. et al. Calcium-dependent plasma membrane binding and cell lysis by perforin are mediated through its C2 domain: a critical role for aspartate residues 429, 435, 483, and 485 but not 491. J. Biol. Chem. 280, 8426–8434 (2005).
Connell, S. D., Heath, G., Olmsted, P. D. & Kisil, A. Critical point fluctuations in supported lipid membranes. Faraday Discuss. 161, 91–111 (2013).
Antia, R., Schlegel, R. A. & Williamson, P. Binding of perforin to membranes is sensitive to lipid spacing and not headgroup. Immunol. Lett. 32, 153–157 (1992).
Hotze, E. M. et al. Monomer–monomer interactions propagate structural transitions necessary for pore formation by the cholesterol-dependent cytolysins. J. Biol. Chem. 287, 24534–24543 (2012).
Traore, D. A. K. et al. Defining the interaction of perforin with calcium and the phospholipid membrane. Biochem. J. 456, 323–335 (2013).
Salvador-Gallego, R. et al. Bax assembly into rings and arcs in apoptotic mitochondria is linked to membrane pores. EMBO J. 35, 389–401 (2016).
Bavdek, A. et al. pH dependence of listeriolysin O aggregation and pore-forming ability. FEBS J. 279, 126–141 (2012).
Jenkins, M. R. et al. Failed CTL/NK cell killing and cytokine hypersecretion are directly linked through prolonged synapse time. J. Exp. Med. 212, 307–317 (2015).
Dombkowski, A. A. Disulfide by design: a computational method for the rational design of disulfide bonds in proteins. Bioinformatics 19, 1852–1853 (2003).
Voskoboinik, I. et al. The functional basis for hemophagocytic lymphohistiocytosis in a patient with co-inherited missense mutations in the perforin (PFN1) gene. J. Exp. Med. 200, 811–816 (2004).
Brennan, A. J. et al. Protection from endogenous perforin: glycans and the C terminus regulate exocytic trafficking in cytotoxic lymphocytes. Immunity 34, 879–892 (2011).
Wessel, D. & Flügge, U. I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 (1984).
Hope, M. J., Bally, M. B., Webb, G. & Cullis, P. R. Production of large unilamellar vesicles by a rapid extrusion procedure—characterization of size distribution, trapped volume and ability to maintain a membrane-potential. Biochim. Biophys. Acta 812, 55–65 (1985).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Dang, T. X., Hotze, E. M., Rouiller, I., Tweten, R. K. & Wilson-Kubalek, E. M. Prepore to pore transition of a cholesterol-dependent cytolysin visualized by electron microscopy. J. Struct. Biol. 150, 100–108 (2005).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
van Heel, M., Harauz, G., Orlova, E. V., Schmidt, R. & Schatz, M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 (1996).
Voskoboinik, I. et al. Perforin activity and immune homeostasis: the common A91V polymorphism in perforin results in both presynaptic and postsynaptic defects in function. Blood 110, 1184–1190 (2007).
Phair, R. D., Gorski, S. A. & Misteli, T. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 375, 393–414 (2004).
Soumpasis, D. M. Theoretical analysis of fluorescence photobleaching recovery experiments. Biophys. J. 41, 95–97 (1983).
Ratto, T. V. & Longo, M. L. Obstructed diffusion in phase-separated supported lipid bilayers: a combined atomic force microscopy and fluorescence recovery after photobleaching approach. Biophys. J. 83, 3380–3392 (2002).
Sackmann, E. Supported membranes: scientific and practical applications. Science 271, 43–48 (1996).
Acknowledgements
This work was funded by the BBSRC (BB/J005932/1, BB/J006254/1 and BB/N015487/1), the ERC (advanced grant no. 294408), the Wellcome Trust (no. 079605/2/06/02), NHMRC Fellowship (1059126), Project (1062706) and Program (1013667) grants, and the Sackler Foundation. The authors thank N. Nand Gosvami and J. Pegman for assistance with early experiments and analysis, and R. Thorogate, A. Ciccone and S. Verschoor for technical support.
Author information
Authors and Affiliations
Contributions
C.L. designed, performed and analysed AFM experiments and co-wrote the paper. A.W.H. designed, performed and analysed AFM and FRAP experiments and co-wrote the paper. A.J.B. expressed mutant perforin, developed the TMH1-lock mutant and carried out haemolysis experiments. N.L. carried out and analysed electron microscopy experiments and co-wrote the paper. S.T. performed and analysed haemolysis experiments. C.M.H. performed mass spectrometric analysis and co-wrote the paper. S.C.K., J.C.W. and M.A.D. contributed to the TMH1-lock mutant design. J.A.T. designed in vitro experiments. I.V., H.R.S. and B.W.H. analysed the data, led the research and co-wrote the paper. All authors read and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 18815 kb)
Supplementary information
Supplementary Movie 1 (MOV 2264 kb)
Supplementary information
Supplementary Movie 2 (MOV 733 kb)
Supplementary information
Supplementary Movie 3 (MOV 489 kb)
Supplementary information
Supplementary Movie 4 (MOV 358 kb)
Supplementary information
Supplementary Movie 5 (MOV 412 kb)
Supplementary information
Supplementary Movie 6 (MOV 506 kb)
Rights and permissions
About this article
Cite this article
Leung, C., Hodel, A., Brennan, A. et al. Real-time visualization of perforin nanopore assembly. Nature Nanotech 12, 467–473 (2017). https://doi.org/10.1038/nnano.2016.303
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2016.303
This article is cited by
-
Mesoporous nanoperforators as membranolytic agents via nano- and molecular-scale multi-patterning
Nature Communications (2024)
-
Cell softness renders cytotoxic T lymphocytes and T leukemic cells resistant to perforin-mediated killing
Nature Communications (2024)
-
A transistor-like pH-sensitive nanodetergent for selective cancer therapy
Nature Nanotechnology (2022)
-
Perforin-2 clockwise hand-over-hand pre-pore to pore transition mechanism
Nature Communications (2022)
-
Applications of atomic force microscopy in immunology
Frontiers of Medicine (2021)