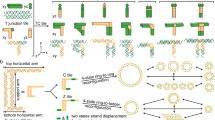
Abstract
Within cells, nanostructures are often organized using local assembly rules that produce long-range order1,2. Because these rules can take into account the cell's current structure and state, they can enable complexes, organelles or cytoskeletal structures to assemble around existing cellular components to form architectures3,4,5,6. Although many methods for self-assembling biomolecular nanostructures have been developed7,8,9,10,11, few can be programmed to assemble structures whose form depends on the identity and organization of structures already present in the environment. Here, we demonstrate that DNA nanotubes can grow to connect pairs of molecular landmarks with different separation distances and relative orientations. DNA tile nanotubes nucleate at these landmarks and grow while their free ends diffuse. The nanotubes can then join end to end to form stable connections, with unconnected nanotubes selectively melted away. Connections form between landmark pairs separated by 1–10 µm in more than 75% of cases and can span a surface or three dimensions. This point-to-point assembly process illustrates how self-assembly kinetics can be designed to produce structures with a desired physical property rather than a specific shape.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Fletcher, D. A. & Mullins, D. Cell mechanics and the cytoskeleton. Nature 463, 485–492 (2010).
Misteli, T. The concept of self-organization in cellular architecture. J. Cell Biol. 155, 181–185 (2001).
Vignaud, T., Blanchoin, L. & Thery, M. Directed cytoskeleton self-organization. Trends Cell Biol. 22, 671–682 (2012).
Wang, N., Tytell, J. D. & Ingber, D. E. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 10, 75–82 (2009).
Greenfield, D. et al. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 7, e1000137 (2009).
Alberts, B. et al. Molecular Biology of the Cell 4th edn (Garland Science, 2002).
Hartgerink, J. D., Beniash, E. & Stupp, S. I. Self-assembly and mineralization of peptide-amphiphile nanofibers. Science 294, 1684–1688 (2001).
Percec, V. et al. Self-assembly of janus dendrimers into uniform dendrimersomes and other complex architectures. Science 328, 1009–1014 (2010).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Barish, R. D., Schulman, R., Rothemund, P. W. K. & Winfree, E. An information-bearing seed for nucleating algorithmic self-assembly. Proc. Natl Acad. Sci. USA 106, 6054–6059 (2009).
Wei, B., Dai, M. J. & Yin, P. Complex shapes self-assembled from single-stranded DNA tiles. Nature 485, 623–626 (2012).
Dinarina, A. et al. Chromatin shapes the mitotic spindle. Cell 138, 502–513 (2009).
Gadde, S. & Heald, R. Mechanisms and molecules of the mitotic spindle. Curr. Biol. 14, R797–R805 (2004).
Helmke, K. J., Heald, R. & Wilbur, J. D. Interplay between spindle architecture and function. Int. Rev. Cell Mol. Biol. 306, 83–125 (2013).
Rothemund, P. W. K. et al. Design and characterization of programmable DNA nanotubes. J. Am. Chem. Soc. 126, 16344–16352 (2004).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Mohammed, A. M. & Schulman, R. Directing self-assembly of DNA nanotubes using programmable seeds. Nano Lett. 13, 4006–4013 (2013).
Rothwell, S. W., Grasser, W. A. & Murphy, D. B. End-to-end annealing of microtubules in vitro. J. Cell Biol. 102, 619–627 (1986).
Ekani-Nkodo, A., Kumar, A. & Fygenson, D. K. Joining and scission in the self-assembly of nanotubes from DNA tiles. Phys. Rev. Lett. 93, 268301 (2004).
Doi, M. & Edwards, S. F. The Theory of Polymer Dynamics (Clarendon, 1988).
Hariadi, R. F., Yurke, B. & Winfree, E. Thermodynamics and kinetics of DNA nanotube polymerization from single-filament measurements. Chem. Sci. 6, 2252–2267 (2015).
Mardanlou, V. et al. A coarse-grained model of DNA nanotube population growth. in DNA Computing and Molecular Programming (eds Rondelez, Y. & Woods, D.) 135–147 (2016).
Brackley, C. A., Morozov, A. N. & Marenduzzo, D. Models for twistable elastic polymers in Brownian dynamics, and their implementation for LAMMPS. J. Chem. Phys. 140, 135103 (2014).
Schiffels, D., Liedl, T. & Fygenson, D. K. Nanoscale structure and microscale stiffness of DNA nanotubes. ACS Nano 7, 6700–6710 (2013).
Zhang, D., Hariadi, R. F., Choi, H. & Winfree, E. Integrating DNA strand-displacement circuitry with DNA tile self-assembly. Nat. Commun. 4, 1965 (2013).
Montagne, K., Plasson, R., Sakai, Y., Fujii, T. & Rondelez, Y. Programming an in vitro DNA oscillator using a molecular networking strategy. Mol. Syst. Biol. 7, 466 (2011).
Tan, S., Campolongo, M., Luo, D. & Cheng, W. Building plasmonic nanostructures with DNA. Nat. Nanotech. 6, 268–276 (2011).
Knudsen, J. B. et al. Routing of individual polymers in designed patterns. Nat. Nanotech. 10, 892–898 (2015).
Wilner, O. I. & Willner, I. Functionalized DNA nanostructures. Chem. Rev. 112, 2528–2556 (2012).
Acknowledgements
The authors thank D. Fygenson, M. Bevan, D. Gracias, E. Winfree, D. Agrawal, S. Schaffter and E. Franco for discussions and advice on the manuscript, J. Liphardt for the use of equipment and advice, and J. Fern, E. Pryce, R. Zuckermann and C. Ajo-Franklin for technical advice. This research has been supported by DOE grant DE-SC0010595, which provided money for materials, supplies and computing time, NSF CAREER award 125387, and the Miller Institute for Basic Science. P.Š. is supported by a grant from the Simons Foundation. Preliminary work related to this project at the Molecular Foundry was supported by the Office of Science, Office of Basic Energy Sciences, of the US Department of Energy, under contract no. DE-AC02-05CH11231.
Author information
Authors and Affiliations
Contributions
A.M.M. and R.S. designed the experiments and carried out the experimental analysis. A.M.M. conducted the experiments. P.Š. and R.S. designed the simulations. P.Š. and J.Z. developed simulations and analysed simulation results. All the authors discussed the results and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 14042 kb)
Supplementary Movie 1
Supplementary Movie 1 (MOV 65 kb)
Supplementary Movie 2
Supplementary Movie 2 (MOV 2871 kb)
Supplementary Movie 3
Supplementary Movie 3 (MOV 27 kb)
Supplementary Movie 4
Supplementary Movie 4 (MOV 118 kb)
Supplementary Movie 5
Supplementary Movie 5 (MOV 515 kb)
Rights and permissions
About this article
Cite this article
Mohammed, A., Šulc, P., Zenk, J. et al. Self-assembling DNA nanotubes to connect molecular landmarks. Nature Nanotech 12, 312–316 (2017). https://doi.org/10.1038/nnano.2016.277
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2016.277
This article is cited by
-
Functional DNA-based cytoskeletons for synthetic cells
Nature Chemistry (2022)
-
Growth and site-specific organization of micron-scale biomolecular devices on living mammalian cells
Nature Communications (2021)
-
A synthetic tubular molecular transport system
Nature Communications (2021)
-
Feedback regulation of crystal growth by buffering monomer concentration
Nature Communications (2020)
-
Towards Active Self-Assembly Through DNA Nanotechnology
Topics in Current Chemistry (2020)