Abstract
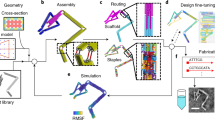
DNA can be programmed to assemble into a variety of shapes and patterns on the nanoscale1,2,3,4,5 and can act as a template for hybrid nanostructures6 such as conducting wires7,8,9, protein arrays8 and field-effect transistors10,11. Current DNA nanostructures are typically in the sub-micrometre range, limited by the sequence space and length of the assembled strands. Here we show that on a patterned biochip12, DNA chains collapse into one-dimensional (1D) fibres that are 20 nm wide and around 70 µm long, each comprising approximately 35 co-aligned chains at its cross-section. Electron beam writing on a photocleavable monolayer was used to immobilize and pattern the DNA molecules, which condense into 1D bundles in the presence of spermidine. DNA condensation can propagate and split at junctions, cross gaps and create domain walls between counterpropagating fronts. This system is inherently adept at solving probabilistic problems and was used to find the possible paths through a maze and to evaluate stochastic switching circuits. This technique could be used to propagate biological or ionic signals13 in combination with sequence-specific DNA nanotechnology or for gene expression in cell-free DNA compartments14.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Seeman, N. C. Nanomaterials based on DNA. Annu. Rev. Biochem. 79, 65–87 (2010).
Jones, M. R., Seeman, N. C. & Mirkin, C. A. Programmable materials and the nature of the DNA bond. Science 347, 1260901 (2015).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Ke, Y. et al. DNA brick crystals with prescribed depths. Nature Chem. 6, 994–1002 (2014).
Catherall, T., Huskisson, D., McAdams, S. & Vijayaraghavan, A. Self-assembly of one dimensional DNA-templated structures. J. Mater. Chem. C 2, 6895–6920 (2014).
Bui, H. et al. Programmable periodicity of quantum dot arrays with DNA origami nanotubes. Nano Lett. 10, 3367–3372 (2010).
Braun, E., Eichen, Y., Sivan, U. & Ben-Yoseph, G. DNA-templated assembly and electrode attachment of a conducting silver wire. Nature 391, 775–778 (1998).
Yan, H., Park, S. H., Finkelstein, G., Reif, J. H. & LaBean, T. H. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science 301, 1882–1884 (2003).
Liu, J. et al. Fabrication of DNA-templated Te and Bi2Te3 nanowires by galvanic displacement. Langmuir 29, 11176–11184 (2013).
Keren, K., Berman, R. S., Buchstab, E., Sivan, U. & Braun, E. DNA-templated carbon nanotube field-effect transistor. Science 302, 1380–1382 (2003).
Maune, H. T. et al. Self-assembly of carbon nanotubes into two-dimensional geometries using DNA origami templates. Nature Nanotech. 5, 61–66 (2009).
Heyman, Y., Buxboim, A., Wolf, S. G., Daube, S. S. & Bar-Ziv, R. H. Cell-free protein synthesis and assembly on a biochip. Nature Nanotech. 7, 374–378 (2012).
Berson, J. et al. Single-layer ionic conduction on carboxyl-terminated silane monolayers patterned by constructive lithography. Nature Mater. 14, 613–621 (2015).
Karzbrun, E., Tayar, A. M., Noireaux, V. & Bar-Ziv, R. H. Programmable on-chip DNA compartments as artificial cells. Science 345, 829–832 (2014).
Hud, N. V. & Vilfan, I. D. Toroidal DNA condensates: unraveling the fine structure and the role of nucleation in determining size. Annu. Rev. Biophys. Biomol. Struct. 34, 295–318 (2005).
Pelta, J. Jr, Durand, D., Doucet, J. & Livolant, F. DNA mesophases induced by spermidine: structural properties and biological implications. Biophys. J. 71, 48–63 (1996).
Livolant, F. Ordered phases of DNA in vivo and in vitro. Phys. A 176, 117–137 (1991).
Yoshikawa, K., Kidoaki, S., Takahashi, M., Vasilevskaya, V. V. & Khokhlov, A. R. Marked discreteness on the coil-globule transition of single duplex DNA. Berich. Bunsen. Gesell. 100, 876–880 (1996).
Iwataki, T., Kidoaki, S., Sakaue, T., Yoshikawa, K. & Abramchuk, S. S. Competition between compaction of single chains and bundling of multiple chains in giant DNA molecules. J. Chem. Phys. 120, 4004–4011 (2004).
Mel'nikov, S. M., Sergeyev, V. G. & Yoshikawa, K. Discrete coil-globule transition of large DNA induced by cationic surfactant. J. Am. Chem. Soc. 117, 2401–2408 (1995).
Petrov, A. S. & Harvey, S. C. Packaging double-helical DNA into viral capsids: structures, forces, and energetics. Biophys. J. 95, 497–502 (2008).
Koltover, I., Wagner, K. & Safinya, C. R. DNA condensation in two dimensions. Proc. Natl Acad. Sci. USA 97, 14046–14051 (2000).
Bracha, D. & Bar-Ziv, R. H. Dendritic and nanowire assemblies of condensed DNA polymer brushes. J. Am. Chem. Soc. 136, 4945–4953 (2014).
Buxboim, A. et al. A single-step photolithographic interface for cell-free gene expression and active biochips. Small 3, 500–510 (2007).
Buxboim, A., Daube, S. S. & Bar-Ziv, R. H. Synthetic gene brushes: a structure-function relationship. Mol. Syst. Biol. 4, 181 (2008).
Bracha, D., Karzbrun, E., Daube, S. S. & Bar-Ziv, R. H. Emergent properties of dense DNA phases toward artificial biosystems on a surface. Acc. Chem. Res. 47, 1912–1921 (2014).
Kolodziej, C. M. & Maynard, H. D. Electron-beam lithography for patterning biomolecules at the micron and nanometer scale. Chem. Mater. 24, 774–780 (2012).
Bracha, D., Karzbrun, E., Shemer, G., Pincus, P. A. & Bar-Ziv, R. H. Entropy-driven collective interactions in DNA brushes on a biochip. Proc. Natl Acad. Sci. USA 110, 4534–4538 (2013).
Hud, N. V. & Downing, K. H. Cryoelectron microscopy of lambda phage DNA condensates in vitreous ice: the fine structure of DNA toroids. Proc. Natl Acad. Sci. USA 98, 14925–14930 (2001).
Cohen, H. et al. Polarizability of G4-DNA observed by electrostatic force microscopy measurements. Nano Lett. 7, 981986 (2007).
Wilhelm, D. & Bruck, J. Stochastic switching circuit synthesis. In 2008 IEEE International Symposium on Information Theory 1388–1392 (IEEE, 2008).
Tsumoto, K., Luckel, F. & Yoshikawa, K. Giant DNA molecules exhibit on/off switching of transcriptional activity through conformational transition. Biophys. Chem. 106, 23–29 (2003).
Estévez-Torres, A. et al. Sequence-independent and reversible photocontrol of transcription/expression systems using a photosensitive nucleic acid binder. Proc. Natl Acad. Sci. USA 106, 12219–12223 (2009).
Acknowledgements
We gratefully acknowledge financial support by the Volkswagen Stiftung (G.P., F.C.S. and R.H.B.-Z., grant no. 86 395), the Israel Science Foundation (R.H.B.-Z.) and the Minerva Foundation (R.H.B.-Z).
Author information
Authors and Affiliations
Contributions
G.P., D.B., F.C.S. and R.H.B.-Z. planned and designed the experiments. G.P., D.B. and O.V. performed the experiments. G.P., D.B., S.S.D., F.C.S. and R.H.B.-Z. wrote the paper; all authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 4548 kb)
Supplementary Movie 1
Supplementary Movie 1 (AVI 626 kb)
Supplementary Movie 2
Supplementary Movie 2 (AVI 1938 kb)
Supplementary Movie 3
Supplementary Movie 3 (AVI 20174 kb)
Rights and permissions
About this article
Cite this article
Pardatscher, G., Bracha, D., Daube, S. et al. DNA condensation in one dimension. Nature Nanotech 11, 1076–1081 (2016). https://doi.org/10.1038/nnano.2016.142
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2016.142
This article is cited by
-
Programming DNA origami patterning with non-canonical DNA-based metallization reactions
Nature Communications (2019)