Abstract
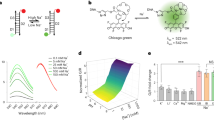
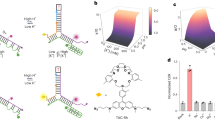
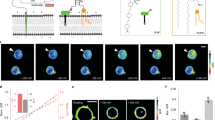
The concentration of chloride ions in the cytoplasm and subcellular organelles of living cells spans a wide range (5–130 mM), and is tightly regulated by intracellular chloride channels or transporters. Chloride-sensitive protein reporters have been used to study the role of these chloride regulators, but they are limited to a small range of chloride concentrations and are pH-sensitive. Here, we show that a DNA nanodevice can precisely measure the activity and location of subcellular chloride channels and transporters in living cells in a pH-independent manner. The DNA nanodevice, called Clensor, is composed of sensing, normalizing and targeting modules, and is designed to localize within organelles along the endolysosomal pathway. It allows fluorescent, ratiometric sensing of chloride ions across the entire physiological regime. We used Clensor to quantitate the resting chloride concentration in the lumen of acidic organelles in Drosophila melanogaster. We showed that lumenal lysosomal chloride, which is implicated in various lysosomal storage diseases, is regulated by the intracellular chloride transporter DmClC-b.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Sheppard, D. N. & Welsh, M. J. Structure and function of the CFTR chloride channel. Physiol. Rev. 79, S23–S45 (1999).
Stauber, T., Weinert, S. & Jentsch, T. J. Cell biology and physiology of CLC chloride channels and transporters. Compr. Physiol. 2, 1701–1744 (2012).
Stauber, T. & Jentsch, T. J. Chloride in vesicular trafficking and function. Annu. Rev. Physiol. 75, 453–477 (2013).
Kuner, T. & Augustine, G. J. A genetically encoded ratiometric indicator for chloride: capturing chloride transients in cultured hippocampal neurons. Neuron 27, 447–459 (2000).
Markova, O., Mukhtarov, M., Real, E., Jacob, Y. & Bregestovski, P. Genetically encoded chloride indicator with improved sensitivity. J. Neurosci. Methods 170, 67–76 (2008).
Arosio, D. et al. Simultaneous intracellular chloride and pH measurements using a GFP-based sensor. Nature Methods 7, 516–518 (2010).
Casey, J. R., Grinstein, S. & Orlowski, J. Sensors and regulators of intracellular pH. Nature Rev. Mol. Cell Biol. 11, 50–61 (2010).
Sonawane, N. D., Thiagarajah, J. R. & Verkman, A. S. Chloride concentration in endosomes measured using a ratioable fluorescent Cl− indicator: evidence for chloride accumulation during acidification. J. Biol. Chem. 277, 5506–5513 (2002).
Krapf, R., Illsley, N. P., Tseng, H. C. & Verkman, A. S. Structure–activity relationships of chloride-sensitive fluorescent indicators for biological application. Anal. Biochem. 169, 142–150 (1988).
Verkman, A. S. Development and biological applications of chloride-sensitive fluorescent indicators. Am. J. Physiol. 259, C375–C388 (1990).
Biwersi, J., Tulk, B. & Verkman, A. S. Long-wavelength chloride-sensitive fluorescent indicators. Anal. Biochem. 219, 139–143 (1994).
Geddes, C. D. Optical halide sensing using fluorescence quenching: theory, simulations and applications—a review. Meas. Sci. Technol. 12, R53 (2001).
Sonawane, N. D. & Verkman, A. S. Determinants of [Cl–] in recycling and late endosomes and Golgi complex measured using fluorescent ligands. J. Cell Biol. 160, 1129–1138 (2003).
Bhatia, D., Sharma, S. & Krishnan, Y. Synthetic, biofunctional nucleic acid-based molecular devices. Curr. Opin. Biotechnol. 22, 475–484 (2011).
Krishnan, Y. & Bathe, M. Designer nucleic acids to probe and program the cell. Trends Cell Biol. 22, 624–633 (2012).
Modi, S. et al. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nature Nanotech. 4, 325–330 (2009).
Modi, S., Nizak, C., Surana, S., Halder, S. & Krishnan, Y. Two DNA nanomachines map pH changes along intersecting endocytic pathways inside the same cell. Nature Nanotech. 8, 459–467 (2013).
Bhatia, D. et al. Icosahedral DNA nanocapsules by modular assembly. Angew. Chem. Int. Ed. 48, 4134–4137 (2009).
Lee, H. et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nature Nanotech. 7, 389–393 (2012).
Surana, S., Bhat, J. M., Koushika, S. P. & Krishnan, Y. An autonomous DNA nanomachine maps spatiotemporal pH changes in a multicellular living organism. Nature Commun. 2, 340 (2011).
Modi, S., Halder, S., Nizak, C. & Krishnan, Y. Recombinant antibody mediated delivery of organelle-specific DNA pH sensors along endocytic pathways. Nanoscale 6, 1144–1152 (2014).
Wilner, S. E. et al. An RNA alternative to human transferrin: a new tool for targeting human cells. Mol. Ther. Nucleic Acids 1, e21 (2012).
Guha, A., Sriram, V., Krishnan, K. S. & Mayor, S. Shibire mutations reveal distinct dynamin-independent and -dependent endocytic pathways in primary cultures of Drosophila hemocytes. J. Cell Sci. 116, 3373–3386 (2003).
Mayle, K. M., Le, A. M. & Kamei, D. T. The intracellular trafficking pathway of transferrin. Biochim. Biophys. Acta 1820, 264–281 (2012).
Bhatia, D., Surana, S., Chakraborty, S., Koushika, S. P. & Krishnan, Y. A synthetic icosahedral DNA-based host–cargo complex for functional in vivo imaging. Nature Commun. 2, 339 (2011).
Huotari, J. & Helenius, A. Endosome maturation. EMBO J. 30, 3481–3500 (2011).
Gupta, G. D. et al. Analysis of endocytic pathways in Drosophila cells reveals a conserved role for GBF1 in internalization via GEECs. PLoS ONE 4, e6768 (2009).
Krapf, R., Berry, C. A. & Verkman, A. S. Estimation of intracellular chloride activity in isolated perfused rabbit proximal convoluted tubules using a fluorescent indicator. Biophys J. 53, 955–962 (1988).
Weinert, S. et al. Lysosomal pathology and osteopetrosis upon loss of H+-driven lysosomal Cl– accumulation. Science 328, 1401–1403 (2010).
Hara-Chikuma, M. et al. ClC-3 chloride channels facilitate endosomal acidification and chloride accumulation. J. Biol. Chem. 280, 1241–1247 (2005).
Hara-Chikuma, M., Wang, Y., Guggino, S. E., Guggino, W. B. & Verkman, A. S. Impaired acidification in early endosomes of ClC-5 deficient proximal tubule. Biochem. Biochem. Biophys. Res. Commun. 329, 941–946 (2005).
Mohammad-Panah, R. et al. An essential role for ClC-4 in transferrin receptor function revealed in studies of fibroblasts derived from Clcn4-null mice. J. Cell Sci. 122, 1229–1237 (2009).
Poët, M. et al. Lysosomal storage disease upon disruption of the neuronal chloride transport protein ClC-6. Proc. Natl Acad. Sci. USA 103, 13854–13859 (2006).
Kasper, D. et al. Loss of the chloride channel ClC-7 leads to lysosomal storage disease and neurodegeneration. EMBO J. 24, 1079–1091 (2005).
Mindell, J. A. Lysosomal acidification mechanisms. Annu. Rev. Physiol. 74, 69–86 (2012).
DiCiccio, J. E. & Steinberg, B. E. Lysosomal pH and analysis of the counter ion pathways that support acidification. J. Gen. Physiol. 137, 385–390 (2011).
Kornak, U. et al. Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell 104, 205–215 (2001).
Scott, C. C. & Gruenberg, J. Ion flux and the function of endosomes and lysosomes: pH is just the start. BioEssays 33, 103–110 (2011).
Weisz, O. A. Acidification and protein traffic. Int. Rev. Cytol. 226, 259–319 (2003).
Sriram, V., Krishnan, K. S. & Mayor, S. Deep-orange and carnation define distinct stages in late endosomal biogenesis in Drosophila melanogaster. J. Cell Biol. 161, 593–607 (2003).
Clemens, J. C. et al. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl Acad. Sci. USA 97, 6499–6503 (2000).
Acknowledgements
The authors thank the Central Imaging and Flow Facility at the National Centre for Biological Sciences (NCBS) for imaging. This work was funded by the Wellcome Trust Department of Biotechnology (DBT), the India Alliance and the University of Chicago. S.S., S.H. and V.P. acknowledge the Council of Scientific and Industrial Research (CSIR), Government of India, for fellowship. FLIM experiments were performed at the Northwestern University Center for Advanced Microscopy supported by NCI CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center.
Author information
Authors and Affiliations
Contributions
S.S., K.C., V.P. and Y.K. conceived and designed the experiments. S.S., V.P. and K.C. performed the experiments. S.H. designed the I4LYA488/A647 used herein. S.S., V.P., K.C. and Y.K. analysed the data. S.S. and Y.K. co-wrote the paper. All authors commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Information (PDF 1979 kb)
Rights and permissions
About this article
Cite this article
Saha, S., Prakash, V., Halder, S. et al. A pH-independent DNA nanodevice for quantifying chloride transport in organelles of living cells. Nature Nanotech 10, 645–651 (2015). https://doi.org/10.1038/nnano.2015.130
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2015.130
This article is cited by
-
Detecting organelle-specific activity of potassium channels with a DNA nanodevice
Nature Biotechnology (2023)
-
A DNA nanodevice for mapping sodium at single-organelle resolution
Nature Biotechnology (2023)
-
A temporally resolved DNA framework state machine in living cells
Nature Machine Intelligence (2023)
-
A spatially-controlled DNA triangular prism nanomachine for AND-gated intracellular imaging of ATP in acidic microenvironment
Microchimica Acta (2023)
-
Cell Rover—a miniaturized magnetostrictive antenna for wireless operation inside living cells
Nature Communications (2022)