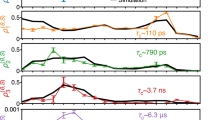
Abstract
For cells to function properly1, membrane proteins must be able to diffuse within biological membranes. The functions of these membrane proteins depend on their position and also on protein–protein and protein–lipid interactions2. However, so far, it has not been possible to study simultaneously the structure and dynamics of biological membranes. Here, we show that the motion of unlabelled membrane proteins can be characterized using high-speed atomic force microscopy3. We find that the molecules of outer membrane protein F (OmpF) are widely distributed in the membrane as a result of diffusion-limited aggregation, and while the overall protein motion scales roughly with the local density of proteins in the membrane, individual protein molecules can also diffuse freely or become trapped by protein–protein interactions. Using these measurements, and the results of molecular dynamics simulations, we determine an interaction potential map and an interaction pathway for a membrane protein, which should provide new insights into the connection between the structures of individual proteins and the structures and dynamics of supramolecular membranes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Engelman, D. M. Membranes are more mosaic than fluid. Nature 438, 578–580 (2005).
Phillips, R., Ursell, T., Wiggins, P. & Sens, P. Emerging roles for lipids in shaping membrane–protein function. Nature 459, 379–385 (2009).
Ando, T. et al. A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl Acad. Sci. USA 98, 12468–12472 (2001).
Schmidt, T., Schütz, G. J., Baumgartner, W., Gruber, H. J. & Schindler, H. Imaging of single molecule diffusion. Proc. Natl Acad. Sci. USA 93, 2926–2929 (1996).
Rosenbusch, P. Characterization of the major envelope protein from Escherichia coli. J. Biol. Chem. 249, 8019–8029 (1974).
Engel, A., Massalski, A., Schindler, H., Dorset, D. L. & Rosenbusch, J. P. Porin channel triplets merge into single outlets in Escherichia coli outer membranes. Nature 317, 643–645 (1985).
Foulds, J. & Chai, T. J. New major outer membrane proteins found in an Escherichia coli tolF mutant resistant to bacteriophage TuIb. J. Bacteriol. 133, 1478–1483 (1978).
Housden, N. G., Loftus, S. R., Moore, G. R., James, R. & Kleanthous, C. Cell entry mechanism of enzymatic bacterial colicins: porin recruitment and the thermodynamics of receptor binding. Proc. Natl Acad. Sci. USA 102, 13849–13854 (2005).
Yamashita, E., Zhalnina, M. V., Zakharov, S. D., Sharma, O. & Cramer, W. A. Crystal structures of the OmpF porin: function in a colicin translocon. EMBO J. 27, 2171–2180 (2008).
Husain, M., Boudier, T., Paul-Gilloteaux, P., Casuso, I. & Scheuring, S. Software for drift compensation, particle tracking and particle analysis of high-speed atomic force microscopy image series. J. Mol. Recognit. 25, 292–298 (2012).
Smith, T. G. Jr, Lange, G. D. & Marks, W. B. Fractal methods and results in cellular morphology—dimensions, lacunarity and multifractals. J. Neurosci. Methods 69, 123–136 (1996).
Witten, T. A. & Sander, L. M. Diffusion-limited aggregation, a kinetic critical phenomenon. Phys. Rev. Lett. 47, 1400–1403 (1981).
Spector, J. et al. Mobility of BtuB and OmpF in the Escherichia coli outer membrane: implications for dynamic formation of a translocon complex. Biophys. J. 99, 3880–3886 (2010).
Kusumi, A., Sako, Y. & Yamamoto, M. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys. J. 65, 2021–2040 (1993).
Mika, J. T. & Poolman, B. Macromolecule diffusion and confinement in prokaryotic cells. Curr. Opin. Biotechnol. 22, 117–126 (2011).
Anne, P. Voronoi and Voronoi-related tessellations in studies of protein structure and interaction. Curr. Opin. Struct. Biol. 14, 233–241 (2004).
Cowan, S. W. et al. Crystal structures explain functional properties of two E. coli porins. Nature 358, 727–733 (1992).
Hoenger, A., Pages, J. M., Fourel, D. & Engel, A. The orientation of porin OmpF in the outer membrane of Escherichia coli. J. Mol. Biol. 233, 400–413 (1993).
Serge, A., Bertaux, N., Rigneault, H. & Marguet, D. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nature Methods 5, 687–694 (2008).
Marguet, D., Lenne, P. F., Rigneault, H. & He, H. T. Dynamics in the plasma membrane: how to combine fluidity and order. EMBO J. 25, 3446–3457 (2006).
García, R. (ed.) in Amplitude Modulation Atomic Force Microscopy 77–90 (Wiley, 2010).
Uchihashi, T., Iino, R., Ando, T. & Noji, H. High-speed atomic force microscopy reveals rotary catalysis of rotorless F1-ATPase. Science 333, 755–758 (2011).
Osborn, M. J. & Wu, H. C. P. Proteins of the outer-membrane of gram-negative bacteria. Annu. Rev. Microbiol. 34, 369–422 (1980).
De Meyer, F. J-M., Venturoli, M. & Smit, B. Molecular simulations of lipid-mediated protein–protein interactions. Biophys. J. 95, 1851–1865 (2008).
Dix, J. A. & Verkman, A. S. Crowding effects on diffusion in solutions and cells. Annu. Rev. Biophys. 37, 247–263 (2008).
Saffman, P. G. & Delbrück, M. Brownian motion in biological membranes. Proc. Natl Acad. Sci. USA 72, 3111–3113 (1975).
Kusumi, A. et al. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 34, 351–378 (2005).
Schirmer, T., Keller, T. A., Wang, Y. F. & Rosenbusch, J. P. Structural basis for sugar translocation through maltoporin channels at 3.1 Å resolution. Science 267, 512–514 (1995).
Melo, E. & Martins, J. Kinetics of bimolecular reactions in model bilayers and biological membranes. A critical review. Biophys. Chem. 123, 77–94 (2006).
Gambin, Y. et al. Lateral mobility of proteins in liquid membranes revisited. Proc. Natl Acad. Sci. USA 103, 2098–2102 (2006).
Acknowledgements
The authors thank C. Nimigean and F. Rico for critical discussion of the manuscript. The authors also thank P. Abeyrathne for support with the lipopolysaccharide detection. Work in the Scheuring laboratory was supported by the Institut Curie, the Institut National de la Santé et Recherche Médicale (INSERM), the Agence Nationale de la Recherche (ANR) and the City of Paris. Work in the Sturgis laboratory was supported by the Centre National de la Recherche Scientifique (CNRS), the Agence Nationale de la Recherche (ANR), Centre Informatique National de l'Enseignement Superieur (CINES) and Aix-Marseille University. Work in the Stahlberg laboratory was supported by the Swiss National Science Foundation (grant no. 315230_127545, and NCCRs Struct Biol and TransCure) and the Swiss Initiative for Systems Biology (SystemsX.ch).
Author information
Authors and Affiliations
Contributions
I.C. and S.S. conceived the experiments. I.C., M.H., M.C. and J.K. performed experiments. I.C., S.S., M.H., P.P-G., J-P.D. and J.N.S. analysed the data. I.C., S.S., J.N.S. and H.S. co-wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 8995 kb)
Supplementary Movie 1
Supplementary Movie 1 (AVI 6732 kb)
Supplementary Movie 2
Supplementary Movie 2 (AVI 708 kb)
Supplementary Movie 3
Supplementary Movie 3 (AVI 1130 kb)
Supplementary Movie 4
Supplementary Movie 4 (AVI 8812 kb)
Supplementary Movie 5
Supplementary Movie 5 (AVI 974 kb)
Supplementary Movie 6
Supplementary Movie 6 (AVI 277 kb)
Supplementary Movie 7
Supplementary Movie 7 (AVI 287 kb)
Supplementary Movie 8
Supplementary Movie 8 (AVI 120 kb)
Supplementary Movie 9
Supplementary Movie 9 (AVI 11992 kb)
Supplementary Movie 10
Supplementary Movie 10 (AVI 8277 kb)
Rights and permissions
About this article
Cite this article
Casuso, I., Khao, J., Chami, M. et al. Characterization of the motion of membrane proteins using high-speed atomic force microscopy. Nature Nanotech 7, 525–529 (2012). https://doi.org/10.1038/nnano.2012.109
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2012.109
This article is cited by
-
Reply to: Antibiotics and hexagonal order in the bacterial outer membrane
Nature Communications (2023)
-
Physical properties of the bacterial outer membrane
Nature Reviews Microbiology (2022)
-
Seeing the unseen: High-resolution AFM imaging captures antibiotic action in bacterial membranes
Nature Communications (2022)
-
Carbon nanotube porin diffusion in mixed composition supported lipid bilayers
Scientific Reports (2020)
-
Recent advances in bioimaging with high-speed atomic force microscopy
Biophysical Reviews (2020)