Abstract
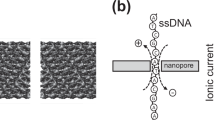
Nanopore analysis is an emerging technique that involves using a voltage to drive molecules through a nanoscale pore in a membrane between two electrolytes, and monitoring how the ionic current through the nanopore changes as single molecules pass through it. This approach allows charged polymers (including single-stranded DNA, double-stranded DNA and RNA) to be analysed with subnanometre resolution and without the need for labels or amplification. Recent advances suggest that nanopore-based sensors could be competitive with other third-generation DNA sequencing technologies, and may be able to rapidly and reliably sequence the human genome for under $1,000. In this article we review the use of nanopore technology in DNA sequencing, genetics and medical diagnostics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Thomas, P. D. & Kejariwal, A. Coding single-nucleotide polymorphisms associated with complex vs. mendelian disease: Evolutionary evidence for differences in molecular effects. Proc. Natl Acad. Sci. USA 101, 15398–15403 (2004).
International HapMap Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005).
Mardis, E. R. Next-generation DNA sequencing methods. Annu. Rev. Genom. Hum. Genet. 9, 387–402 (2008).
Metzker, M. L. Sequencing technologies — the next generation. Nature Rev. Genet. 11, 31–46 (2010).
Coulter, W. H. Means for counting particles suspended in a fluid. US patent 2,656,508 (1953).
Church, G., Deamer, D. W., Branton, D., Baldarelli, R. & Kasianowicz, J. Characterization of individual polymer molecules based on monomer-interface interactions. US patent 5,795,782 (1995).
Deamer, D. W. & Branton, D. Characterization of nucleic acids by nanopore analysis. Acc. Chem. Res. 35, 817–825 (2002).
Rhee, M. & Burns, M. A. Nanopore sequencing technology: research trends and applications. Trends Biotechnol. 24, 580–586 (2006).
Dekker, C. Solid-state nanopores. Nature Nanotech. 2, 209–215 (2007).
Healy, K. Nanopore-based single-molecule DNA analysis. Nanomedicine 2, 459–481 (2007).
Branton, D. et al. The potential and challenges of nanopore sequencing. Nature Biotechnol. 26, 1146–1153 (2008). This review article assesses the feasibility of various nanopore sequencing techniques that are currently under development (both optical and electrical).
Deamer, D. W. Nanopore analysis of nucleic acids bound to exonucleases and polymerases. Annu. Rev. Biophys. 39, 79–90 (2010). This review article provides a historical perspective on the field of nanopore DNA sequencing and elaborates on nanopore-based enzyme-mediated sequencing approaches.
Iqbal, S. & Bashir, R. Nanopores: Sensing and Fundamental Biological Interactions (Springer, 2011).
Kasianowicz, J. J., Brandin, E., Branton, D. & Deamer, D. W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl Acad. Sci. USA 93, 13770–13773 (1996). This is the first experimental report of electronic detection of a DNA molecule passing through a nanopore (α-haemolysin) and marks the start of the nanopore sequencing field.
Akeson, M., Branton, D., Kasianowicz, J. J., Brandin, E. & Deamer, D. W. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys. J. 77, 3227–3233 (1999).
Meller, A. & Branton, D. Single molecule measurements of DNA transport through a nanopore. Electrophoresis 23, 2583–2591 (2002).
Benner, S. et al. Sequence-specific detection of individual DNA polymerase complexes in real time using a nanopore. Nature Nanotech. 2, 718–724 (2007).
Cockroft, S. L., Chu, J., Amorin, M. & Ghadiri, M. R. A single-molecule nanopore device detects DNA polymerase activity with single-nucleotide resolution. J. Am. Chem. Soc. 130, 818–820 (2008).
Lieberman, K. R. et al. Processive replication of single DNA molecules in a nanopore catalyzed by phi29 DNA polymerase. J. Am. Chem. Soc. 132, 17961–17972 (2010). Proof-of-principle experiments demonstrating that a biological nanopore with a coupled polymerase can be used for both strand sequencing and mechanistic studies of enzyme function.
Olasagasti, F. et al. Replication of individual DNA molecules under electronic control using a protein nanopore. Nature Nanotech. 5, 798–806 (2010).
Rincon-Restrepo, M., Mikhailova, E., Bayley, H. & Maglia, G. Controlled translocation of individual DNA molecules through protein nanopores with engineered molecular brakes. Nano Lett. 11, 746–750 (2011).
Mitchell, N. & Howorka, S. Chemical tags facilitate the sensing of individual DNA strands with nanopores. Angew. Chem. Int. Ed. 47, 5565–5568 (2008).
Clarke, J. et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nature Nanotech. 4, 265–270 (2009). Proof-of-principle experiments demonstrating the discrimination of individual mononucleotides using a biological nanopore. Future efforts to couple this with an exonuclease might enable a 'sequencing by digestion' approach.
Stoddart, D., Heron, A. J., Mikhailova, E., Maglia, G. & Bayley, H. Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc. Natl Acad. Sci. USA 106, 7702–7707 (2009).
Faller, M., Niederweis, M. & Schulz, G. E. The structure of a mycobacterial outer-membrane channel. Science 303, 1189–1192 (2004).
Derrington, I. M. et al. Nanopore DNA sequencing with MspA. Proc. Natl Acad. Sci. USA 107, 16060–16065 (2010). Proof-of-principle experiments demonstrating duplex interrupted sequencing using MspA are reported here.
Wendell, D. et al. Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores. Nature Nanotech. 4, 765–772 (2009).
Jing, P., Haque, F., Shu, D., Montemagno, C. & Guo, P. X. One-way traffic of a viral motor channel for double-stranded DNA translocation. Nano Lett. 10, 3620–3627 (2010).
Groves, J. T., Ulman, N. & Boxer, S. G. Micropatterning fluid lipid bilayers on solid supports. Science 275, 651–653 (1997).
Mager, M. D. & Melosh, N. A. Nanopore-spanning lipid bilayers for controlled chemical release. Adv. Mater. 20, 4423–4427 (2008).
White, R. J. et al. Ionic conductivity of the aqueous layer separating a lipid bilayer membrane and a glass support. Langmuir 22, 10777–10783 (2006).
Venkatesan, B. M. et al. Lipid bilayer coated Al2O3 nanopore sensors: towards a hybrid biological solid-state nanopore. Biomed. Microdevices 13, 671–682 (2011).
Chung, M. & Boxer, S. G. Stability of DNA-tethered lipid membranes with mobile tethers. Langmuir 27, 5492–5497 (2011).
Langford, K. W., Penkov, B., Derrington, I. M. & Gundlach, J. H. Unsupported planar lipid membranes formed from mycolic acids of Mycobacterium tuberculosis. J. Lipid Res. 52, 272–277 (2011).
Knoll, W., Köper, I., Naumann, R. & Sinner, E-K. Tethered bimolecular lipid membranes — a novel model membrane platform. Electrochim. Acta 53, 6680–6689 (2008).
Storm, A. J., Chen, J. H., Ling, X. S., Zandbergen, H. W. & Dekker, C. Fabrication of solid-state nanopores with single nanometre precision. Nature Mater. 2, 537–540 (2003).
Venkatesan, B. M. et al. Highly sensitive, mechanically stable nanopore sensors for DNA analysis. Adv. Mater. 21, 2771–2776 (2009).
Kim, M. J., Wanunu, M., Bell, D. C. & Meller, A. Rapid fabrication of uniformly sized nanopores and nanopore arrays for parallel DNA analysis. Adv. Mater. 18, 3149–3153 (2006).
Nam, S-W., Rooks, M. J., Kim, K-B. & Rossnagel, S. M. Ionic field effect transistors with sub-10 nm multiple nanopores. Nano Lett. 9, 2044–2048 (2009).
McNally, B. et al. Optical recognition of converted DNA nucleotides for single-molecule DNA sequencing using nanopore arrays. Nano Lett. 10, 2237–2244 (2010). The use of hybridized fluorescent probes allows optical sequence readout.
Li, J. et al. Ion-beam sculpting at nanometre length scales. Nature 412, 166–169 (2001).
Salisbury, I. G., Timsit, R. S., Berger, S. D. & Humphreys, C. J. Nanometer scale electron beam lithography in inorganic materials. Appl. Phys. Lett. 45, 1289–1291 (1984).
Healy, K., Schiedt, B. & Morrison, A. P. Solid-state nanopore technologies for nanopore-based DNA analysis. Nanomedicine 2, 875–897 (2007).
Venkatesan, B. M., Shah, A. B., Zuo, J. M. & Bashir, R. DNA sensing using nanocrystalline surface-enhanced Al2O3 nanopore sensors. Adv. Funct. Mater. 20, 1266–1275 (2010).
Hoogerheide, D. P., Garaj, S. & Golovchenko, J. A. Probing surface charge fluctuations with solid-state nanopores. Phys. Rev. Lett. 102, 256804 (2009).
George, S. M. Atomic layer deposition: an overview. Chem. Rev. 110, 111–131 (2009).
Geim, A. K. Graphene: status and prospects. Science 324, 1530–1534 (2009).
Fischbein, M. D. & Drndic, M. Electron beam nanosculpting of suspended graphene sheets. Appl. Phys. Lett. 93, 113107–113103 (2008). This is the first report of nanopore fabrication in a suspended graphene membrane using an electron beam and has led to subsequent studies of DNA translocation through graphene nanopores.
Girit, Ç. Ö. et al. Graphene at the edge: stability and dynamics. Science 323, 1705–1708 (2009).
Garaj, S. et al. Graphene as a subnanometre trans-electrode membrane. Nature 467, 190–193 (2010). In addition to containing experimental data showing DNA translocation through a graphene nanopore (see also refs 52 and 53 ), this paper includes a calculation of the spatial resolution that is possible with monolayer graphene nanopore sensors.
Merchant, C. A. et al. DNA translocation through graphene nanopores. Nano Lett. 10, 2915–2921 (2010).
Schneider, G. F. et al. DNA translocation through graphene nanopores. Nano Lett. 10, 3163–3167 (2010).
Hall, J. E. Access resistance of a small circular pore. J. Gen. Physiol. 66, 531–532 (1975).
Song, B. et al. Atomic-scale electron-beam sculpting of near-defect-free graphene nanostructures. Nano Lett. 11, 2247–2250 (2011).
Li, J., Gershow, M., Stein, D., Brandin, E. & Golovchenko, J. A. DNA molecules and configurations in a solid-state nanopore microscope. Nature Mater. 2, 611–615 (2003).
Storm, A. J. et al. Fast DNA translocation through a solid-state nanopore. Nano Lett. 5, 1193–1197 (2005).
Wanunu, M., Morrison, W., Rabin, Y., Grosberg, A. Y. & Meller, A. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nature Nanotech. 5, 160–165 (2010).
Calin, G. A. & Croce, C. M. MicroRNA signatures in human cancers. Nature Rev. Cancer 6, 857–866 (2006).
Volinia, S. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA 103, 2257–2261 (2006).
Wanunu, M. et al. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nature Nanotech. 5, 807–814 (2010).
Strathdee, G. & Brown, R. Aberrant DNA methylation in cancer: potential clinical interventions. Expert Rev. Mol. Med. 4, 1–17 (2002).
Lee, W. H., Isaacs, W. B., Bova, G. S. & Nelson, W. G. CG island methylation changes near the GSTP1 gene in prostatic carcinoma cells detected using the polymerase chain reaction: A new prostate cancer biomarker. Cancer Epidem. Biomar. 6, 443–450 (1997).
Laird, P. W. The power and the promise of DNA methylation markers. Nature Rev. Cancer 3, 253–266 (2003).
Mirsaidov, U. et al. Nanoelectromechanics of methylated DNA in a synthetic nanopore. Biophys. J. 96, L32–L34 (2009).
Wanunu, M. et al. Discrimination of methylcytosine from hydroxymethylcytosine in DNA molecules. J. Am. Chem. Soc. 133, 486–492 (2010).
Botstein, D. & Risch, N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nature Genet. 33, 228–237 (2003).
Zhao, Q. et al. Detecting SNPs using a synthetic nanopore. Nano Lett. 7, 1680–1685 (2007).
Singer, A. et al. Nanopore based sequence specific detection of duplex DNA for genomic profiling. Nano Lett. 10, 738–742 (2010).
Iqbal, S. M., Akin, D. & Bashir, R. Solid-state nanopore channels with DNA selectivity. Nature Nanotech. 2, 243–248 (2007).
Wanunu, M. & Meller, A. Chemically modified solid-state nanopores. Nano Lett. 7, 1580–1585 (2007).
Siwy, Z. S. & Howorka, S. Engineered voltage-responsive nanopores. Chem. Soc. Rev. 39, 1115–1132 (2009).
Kowalczyk, S. W. et al. Single-molecule transport across an individual biomimetic nuclear pore complex. Nature Nanotech. 6, 433–438 (2011).
Yusko, E. C. et al. Controlling protein translocation through nanopores with bio-inspired fluid walls. Nature Nanotech. 6, 253–260 (2011).
Hall, A. R. et al. Hybrid pore formation by directed insertion of alpha-haemolysin into solid-state nanopores. Nature Nanotech. 5, 874–877 (2010).
Vercoutere, W. et al. Rapid discrimination among individual DNA hairpin molecules at single-nucleotide resolution using an ion channel. Nature Biotechnol. 19, 248–252 (2001).
Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009).
Drmanac, R. et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327, 78–81 (2010).
Rothberg, J. M. et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 475, 348–352 (2011).
Karnik, R., Duan, C., Castelino, K., Daiguji, H. & Majumdar, A. Rectification of ionic current in a nanofluidic diode. Nano Lett. 7, 547–551 (2007).
Jin, X. & Aluru, N. R. Gated transport in nanofluidic devices. Microfluid. Nanofluid. 11, 297–306 (2011).
Liu, H. et al. Translocation of single-stranded DNA through single-walled carbon nanotubes. Science 327, 64–67 (2010).
Nelson, T., Zhang, B. & Prezhdo, O. V. Detection of nucleic acids with graphene nanopores: ab initio characterization of a novel sequencing device. Nano Lett. 10, 3237–3242 (2010).
Min, S. K., Kim, W. Y., Cho, Y. & Kim, K. S. Fast DNA sequencing with a graphene-based nanochannel device. Nature Nanotech. 6, 162–165 (2011).
Postma, H. W. C. Rapid sequencing of individual DNA molecules in graphene nanogaps. Nano Lett. 10, 420–425 (2010).
Prasongkit, J., Grigoriev, A., Pathak, B., Ahuja, R. & Scheicher, R. H. Transverse conductance of DNA nucleotides in a graphene nanogap from first principles. Nano Lett. 11, 1941–1945 (2011).
Luan, B. et al. Base-by-base ratcheting of single stranded DNA through a solid-state nanopore. Phys. Rev. Lett. 104, 238103 (2010). IBM's DNA transistor architecture and proposed approach to nanopore-based single-molecule DNA sequencing are presented here.
Huang, S. et al. Identifying single bases in a DNA oligomer with electron tunnelling. Nature Nanotech. 5, 868–873 (2010).
Zwolak, M. & Di Ventra, M. Physical approaches to DNA sequencing and detection. Rev. Mod. Phys. 80, 141–165 (2008).
Tanaka, H. & Kawai, T. Partial sequencing of a single DNA molecule with a scanning tunnelling microscope. Nature Nanotech. 4, 518–522 (2009).
Tsutsui, M., Taniguchi, M., Yokota, K. & Kawai, T. Identifying single nucleotides by tunnelling current. Nature Nanotech. 5, 286–290 (2010). This is the first report of a nanofabricated gap junction being used to discriminate individual nucleotides through electron tunnelling measurements.
Taniguchi, M., Tsutsui, M., Yokota, K. & Kawai, T. Fabrication of the gating nanopore device. Appl. Phys. Lett. 95, 123701–123703 (2009).
Ivanov, A. P. et al. DNA tunneling detector embedded in a nanopore. Nano Lett. 11, 279–285 (2010).
Asmann, Y. W., Kosari, F., Wang, K., Cheville, J. C. & Vasmatzis, G. Identification of differentially expressed genes in normal and malignant prostate by electronic profiling of expressed sequence tags. Cancer Res. 62, 3308–3314 (2002).
Feldman, A. L. et al. Discovery of recurrent t(6;7)(p25.3;q32.3) translocations in ALK-negative anaplastic large cell lymphomas by massively parallel genomic sequencing. Blood 117, 915–919 (2010).
Ling, X. S., Bready, B. & Pertsinidis, A. Hybridization-assisted nanopore sequencing of nucleic acids. US patent 2007 0190542 (2007). This approach, called HANS, is being commercially developed by NABsys: 6-mer oligonucleotide probes are hybridized to ssDNA and current versus time traces reveal the position of the probe on the ssDNA template.
Astier, Y., Braha, O. & Bayley, H. Toward single molecule DNA sequencing: direct identification of ribonucleoside and deoxyribonucleoside 5′-monophosphates by using an engineered protein nanopore equipped with a molecular adapter. J. Am. Chem. Soc. 128, 1705–1710 (2006).
Lagerqvist, J., Zwolak, M. & Di Ventra, M. Fast DNA sequencing via transverse electronic transport. Nano Lett. 6, 779–782 (2006).
Heng, J. B. et al. Beyond the gene chip. Bell Labs Tech. J. 10, 5–22 (2005).
Gracheva, M. E. et al. Simulation of the electric response of DNA translocation through a semiconductor nanopore-capacitor. Nanotechnology 17, 622–633 (2006).
Sigalov, G., Comer, J., Timp, G. & Aksimentiev, A. Detection of DNA sequences using an alternating electric field in a nanopore capacitor. Nano Lett. 8, 56–63 (2007).
Meller, A., Nivon, L., Brandin, E., Golovchenko, J. & Branton, D. Rapid nanopore discrimination between single polynucleotide molecules. Proc. Natl Acad. Sci. USA 97, 1079–1084 (2000).
Howorka, S., Cheley, S. & Bayley, H. Sequence-specific detection of individual DNA strands using engineered nanopores. Nature Biotechnol. 19, 636–639 (2001).
Bates, M., Burns, M. & Meller, A. Dynamics of DNA molecules in a membrane channel probed by active control techniques. Biophys. J. 84, 2366–2372 (2003).
Borsenberger, V., Mitchell, N. & Howorka, S. Chemically labeled nucleotides and oligonucleotides encode DNA for sensing with nanopores. J. Am. Chem. Soc. 131, 7530–7531 (2009).
Chen, P. et al. Probing single DNA molecule transport using fabricated nanopores. Nano Lett. 4, 2293–2298 (2004).
Storm, A. J., Chen, J. H., Zandbergen, H. W. & Dekker, C. Translocation of double-strand DNA through a silicon oxide nanopore. Phys. Rev. E 71, 051903 (2005).
Fologea, D., Uplinger, J., Thomas, B., McNabb, D. S. & Li, J. Slowing DNA translocation in a solid-state nanopore. Nano Lett. 5, 1734–1737 (2005).
Kim, Y. R. et al. Nanopore sensor for fast label-free detection of short double-stranded DNAs. Biosensors Bioelectron. 22, 2926–2931 (2007).
Wanunu, M., Sutin, J., McNally, B., Chow, A. & Meller, A. DNA translocation governed by interactions with solid-state nanopores. Biophys. J. 95, 4716–4725 (2008).
Chen, Z. et al. DNA translocation through an array of kinked nanopores. Nature Mater. 9, 667–675 (2010).
Acknowledgements
B.M.V. is a trainee supported by the Midwestern Cancer Nanotechnology Training Center (NIH-NCI R25 CA154015). Support from the National Institutes of Health (R21 CA155863) and the National Science Foundation (EEC-0425626) is also acknowledged. The authors thank J. Hanlon-Sinn (Beckman Institute of Advanced Technology, University of Illinois at Urbana-Champaign) for the images in Fig. 6, and M. Drndic (University of Pennsylvania) for valuable discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Venkatesan, B., Bashir, R. Nanopore sensors for nucleic acid analysis. Nature Nanotech 6, 615–624 (2011). https://doi.org/10.1038/nnano.2011.129
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2011.129
This article is cited by
-
TCF7L2 acts as a molecular switch in midbrain to control mammal vocalization through its DNA binding domain but not transcription activation domain
Molecular Psychiatry (2023)
-
Graphene–molecule–graphene single-molecule junctions to detect electronic reactions at the molecular scale
Nature Protocols (2023)
-
Unidirectional single-file transport of full-length proteins through a nanopore
Nature Biotechnology (2023)
-
Adenosine A1 receptor ligands bind to α-synuclein: implications for α-synuclein misfolding and α-synucleinopathy in Parkinson’s disease
Translational Neurodegeneration (2022)
-
Driven translocation of a semiflexible polymer through a conical channel in the presence of attractive surface interactions
Scientific Reports (2022)