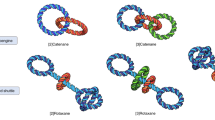
Abstract
Tensegrity, or tensional integrity, is a property of a structure indicating a reliance on a balance between components that are either in pure compression or pure tension for stability1,2. Tensegrity structures exhibit extremely high strength-to-weight ratios and great resilience, and are therefore widely used in engineering, robotics and architecture3,4. Here, we report nanoscale, prestressed, three-dimensional tensegrity structures in which rigid bundles of DNA double helices resist compressive forces exerted by segments of single-stranded DNA that act as tension-bearing cables. Our DNA tensegrity structures can self-assemble against forces up to 14 pN, which is twice the stall force of powerful molecular motors such as kinesin or myosin5,6. The forces generated by this molecular prestressing mechanism can be used to bend the DNA bundles or to actuate the entire structure through enzymatic cleavage at specific sites. In addition to being building blocks for nanostructures, tensile structural elements made of single-stranded DNA could be used to study molecular forces, cellular mechanotransduction and other fundamental biological processes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Snelson, K. Snelson on the tensegrity invention. Int. J. Space Struct. 11, 43–48 (1996).
Fuller, B. Synergetics—Explorations in the Geometry of Thinking Vols I and II (Macmillan Publishing, 1975, 1979).
Skelton, R. E., Adhikari, R., Pinaud, J. P. & Chan, W. An introduction to the mechanics of tensegrity structures. Proceedings of the 40th IEEE Conference on Decision and Control 5, 4254–4259 (2001).
Sultan, C. Corless, M. & Skelton, R. E. The prestressabilty problem of tensegrity structures: some analytical solutions. Int. J. Solids Struct. 38, 5223–5252 (2001).
Visscher, K., Schnitzer, M. J. & Block, S. M. Single kinesin molecules studied with a molecular force clamp. Nature 400, 184–189 (1999).
Clemen, A. E. et al. Force-dependent stepping kinetics of myosin-V. Biophys. J. 88, 4402–4410 (2005).
Ingber, D. E. The architecture of life. Sci. Am. 278, 48–57 (1998).
Ingber, D. E. Cellular mechanotransduction: putting all the pieces together again. FASEB J. 20, 811–827 (2006).
Shih, W. M., Quispe J. D. & Joyce, G. F. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature 427, 618–621 (2004).
Liu, D., Wang, M. S., Deng, Z. X., Walulu, R. & Mao, C. D. Tensegrity: construction of rigid DNA triangles with flexible four-arm DNA junctions. J. Am. Chem. Soc. 126, 2324–2325 (2004).
Zhang, C. et al. Conformational flexibility facilitates self-assembly of complex DNA nanostructures. Proc. Natl Acad. Sci. 31, 10665–10669 (2008).
He, Y. et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature 452, 198–201 (2008).
Zheng, J. et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 461, 74–77 (2009).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 287–302 (2006).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Smith, S. B., Cui, Y. J. & Bustamante, C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science 271, 795–799 (1996).
Douglas, S. M. et al. Rapid prototyping of three-dimensional DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009).
Marko, J. F. & Siggia, E. D. Stretching DNA. Macromolecules 28, 8759–8770 (1995).
Douglas, S. M., Chou, J. J. & Shih, W. M. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Natl Acad. Sci. USA 104, 6644–6648 (2007).
Howard, J. Mechanics of Motor Proteins and the Cytoskeleton (Sinauer Associates, 2001).
Goodman, R. P. et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science 310, 1661–1665 (2005).
Saleh, O. A., McIntosh, D. B., Pincus, P. & Ribeck, N. Nonlinear low-force elasticity of single-stranded DNA molecules. Phys. Rev. Lett. 102, 068301 (2009).
Högberg, B., Liedl, T. & Shih, W. M. Folding DNA origami from a double-stranded source of scaffold. J. Am. Chem. Soc. 131, 91544–94155 (2009).
Ingber, D. E. The origin of cellular life. Bioessays 22, 1160–1167 (2000).
Shroff, H. et al. Biocompatible force sensor with optical readout and dimensions of 6 nm3. Nano Lett. 5, 1509–1514 (2005).
Acknowledgements
The authors thank O. Hallatschek, R. Neher, H. Dietz and S. Douglas for helpful discussions and advice. This work was funded by the Wyss Institute for Biologically Inspired Engineering, and Deutscher Akademischer Austauschdienst (DAAD; T.L.), Swedish Science Council (Vetenskapsrådet) Fellowship (B.H.) and Claudia Adams Barr Program Investigator and NIH New Innovator (1DP2OD004641-01; W.M.S.) grants.
Author information
Authors and Affiliations
Contributions
T.L., D.E.I. and W.M.S. conceived and designed the research. T.L. designed the DNA shapes. T.L., B.H. and J.T. performed the experiments. T.L. and B.H. analysed the data, and T.L., B.H., D.E.I. and W.M.S. co-wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 16607 kb)
Rights and permissions
About this article
Cite this article
Liedl, T., Högberg, B., Tytell, J. et al. Self-assembly of three-dimensional prestressed tensegrity structures from DNA. Nature Nanotech 5, 520–524 (2010). https://doi.org/10.1038/nnano.2010.107
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2010.107
This article is cited by
-
Multiscale tensegrity model for the tensile properties of DNA nanotubes
Applied Mathematics and Mechanics (2023)
-
A modular spring-loaded actuator for mechanical activation of membrane proteins
Nature Communications (2022)
-
Catastrophe in Elastic Tensegrity Frameworks
Arnold Mathematical Journal (2022)
-
The biological applications of DNA nanomaterials: current challenges and future directions
Signal Transduction and Targeted Therapy (2021)
-
Linking Tensegrity to Sports Team Collective Behaviors: Towards the Group-Tensegrity Hypothesis
Sports Medicine - Open (2020)