Abstract
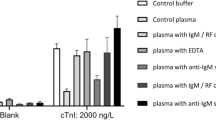
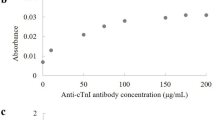
Early detection of the protein marker troponin I in patients with a higher risk of acute myocardial infarction1,2,3,4,5 can reduce the risk of death from heart attacks6,7,8,9,10. Most troponin assays are currently based on the conventional enzyme linked immunosorbent assay and have detection limits in the nano- and picomolar range11. Here, we show that by combining viral nanoparticles, which are engineered to have dual affinity for troponin antibodies and nickel, with three-dimensional nanostructures including nickel nanohairs, we can detect troponin levels in human serum samples that are six to seven orders of magnitude lower than those detectable using conventional enzyme linked immunosorbent assays11,12,13,14,15,16. The viral nanoparticle helps to orient the antibodies for maximum capture of the troponin markers. High densities of antibodies on the surfaces of the nanoparticles and nanohairs lead to greater binding of the troponin markers, which significantly enhances detection sensitivities. The nickel nanohairs are re-useable and can reproducibly differentiate healthy serum from unhealthy ones. We expect other viral nanoparticles to form similar highly sensitive diagnostic assays for a variety of other protein markers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Adams, J. E. et al. Cardiac troponin I. A marker with high specificity for cardiac injury. Circulation 88, 101–106 (1993).
Adams, J. E., Schechtman, K. B., Landt, Y., Ladenson, J. H. & Jaffe, A. S. Comparable detection of acute myocardial infarction by creatine kinase MB isoenzyme and cardiac troponin I. Clin. Chem. 40, 1291–1295 (1994).
Thygesen, K., Alpert, J. S. & White, H. D. Universal definition of myocardial infarction. J. Am. Coll. Cardiol. 50, 2173–2195 (2007).
Morrow, D. A. et al. National Academy of Clinical Biochemistry Laboratory Medicine practice guidelines: clinical characteristics and utilization of biochemical markers in acute coronary syndromes. Clin. Chem. 53, 552–574 (2007).
Gibler, W. B. et al. Practical implementation of the guidelines for unstable angina/non-ST-segment elevation myocardial infarction in the emergency department. Ann. Emerg. Med. 46, 185–197 (2005).
Antman, E. M. et al. Cardiac-specific troponin I levels to predict the risk of mortality in patients with acute coronary syndromes. N. Engl. J. Med. 335, 1342–1349 (1996).
Wu, A. H. B. & Jaffe, A. S. The clinical need for high-sensitivity cardiac troponin assays for acute coronary syndromes and the role for serial testing. Am. Heart J. 155, 208–214 (2008).
Benamer, H. et al. Elevated cardiac troponin I predicts a high-risk angiographic anatomy of the culprit lesion in unstable angina. Am. Heart J. 137, 815–820 (1999).
Heeschen, C., van den Brand, M. J., Hamm, C. W. & Simoons, M. L. Angiographic findings in patients with refractory unstable angina according to troponin T status. Circulation 100, 1509–1514 (1999).
Wong, G. C. et al. Elevations in troponin T and I are associated with abnormal tissue level perfusion: A TACTICS-TIMI 18 substudy. Circulation 106, 202–207 (2002).
Rosi, N. L. & Mirkin, C. A. Nanostructures in biodiagnostics. Chem. Rev. 105, 1547–1562 (2005).
Hirsch, L. R., Jackson, J. B., Lee, A., Halas, N. J. & West, J. A whole blood immunoassay using gold nanoshells. Anal. Chem. 75, 2377–2381 (2003).
Nam, J. M., Park, S. J. & Mirkin, C. A. Bio-barcodes based on oligonucleotide-modified nanoparticles. J. Am. Chem. Soc. 124, 3820–3821 (2002).
Niemeyer, C. M. & Ceyhan, B. DNA-directed functionalization of colloidal gold with proteins. Angew. Chem. Int. Ed. 40, 3685–3688 (2001).
Chien, R. J. et al. Noncovalent functionalization of carbon nanotubes for highly specific electronic biosensors. Proc. Natl Acad. Sci. 100, 4984–4989 (2003).
Wang, J., Polsky, R., Merkoci, A. & Turner, K. L. DNA-based amplified bioelectronic detection and coding of proteins. Angew. Chem. Int. Ed. 43, 2158–2161 (2004).
Rowe, C. A. et al. Array biosensor for simultaneous identification of bacterial, viral and protein analytes. Anal. Chem. 71, 3846–3852 (1999).
Gouda, H. et al. NMR study of the interaction between the B domain of staphylococcal protein A and the Fc portion of immunoglobulin G. Biochemistry 37, 129–136 (1998).
Deisenhofer, J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-Å. resolution. Biochemistry 20, 2361–2370 (1981).
Böttcher, B., Wynne, S. A. & Crowther, R. A. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 386, 88–91 (1997).
Crowther, R. A. et al. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell 77, 943–950 (1994).
Fischlechner, M. & Donath, E. Viruses as building blocks for materials and devices. Angew Chem. Int. Ed. 46, 3184–3193 (2007).
Toellner, L., Fischlechner, M., Ferko, B., Grabherr, R. M. & Donath, E. Virus-coated layer-by-layer colloids as a multiplex suspension array for the detection and quantification of virus-specific antibodies. Clin. Chem. 52, 1575–1583 (2006).
Yang, L. M. C. et al. Virus electrodes for universal biodetection. Anal. Chem. 78, 3265–3270 (2006).
Willis, S. et al. Virus-like particles as quantitative probes of membrane protein interactions. Biochemistry 47, 6988–6990 (2008).
Datta, M. et al. High relaxivity gadolinium hydroxypyridonate-viral capsid conjugates: nanosized MRI contrast agents. J. Am. Chem. Soc. 130, 2546–2552 (2008).
Fischlechner, M. et al. Fusion of enveloped virus nanoparticles with polyelectrolyte-supported lipid membranes for the design of bio/nonbio interfaces. Nano Lett. 7, 3540–3546 (2007).
Apple, F. S., Smith, S. W., Pearce, L. A., Ler, R. & Murakami, M. M. Use of the centaur TnI-ultra assay for detection of myocardial infarction and adverse events in patients presenting with symptoms suggestive of acute coronary syndrome. Clin. Chem. 54, 723–728 (2008).
Ahn, J. Y. et al. Heterologous gene expression using self-assembled supra-molecules with high affinity for HSP70 chaperone. Nucl. Acids Res. 33, 3751–3762 (2005).
Acknowledgements
This study was supported by the National Research Laboratory Project (a main project that supported this work) of the Ministry of Education, Science and Technology (grant no. ROA-2007-000-20084-0), the Korea Health 21 R&D Project of the Ministry of Health, Welfare and Family Affairs of the Republic of Korea (grant no. A050750), the Pioneer Research Center Program (grant no. M10711160001-08M1116-00110), the Microbial Genomics and Applications Center at KRIBB, the Seoul R&BD program (no. 10920), and the Korea Research Foundation (grant no. KRF-2006-005-J03603).
Author information
Authors and Affiliations
Contributions
J.L. conceived the viral particle-based assay experiments, conducted the data analysis and wrote the paper. Y.K.K. conceived the nanohair concept, conducted the data analysis and co-wrote the paper. J.S.P. and E.J.L. performed the viral particle synthesis and assay experiments and collected the data. M.K.C. performed the nanohair synthesis and SEM analysis. J.S.P., E.J.L. and M.K.C. contributed equally to this work. K.Y.A. analysed assay data. K.E.L. and S.-S.H. contributed TEM analysis tools. J.H.J. and Y.C. contributed human sera for diagnostic assay.
Corresponding authors
Supplementary information
Supplementary Information
Supplementary Information (PDF 965 kb)
Rights and permissions
About this article
Cite this article
Park, JS., Cho, M., Lee, E. et al. A highly sensitive and selective diagnostic assay based on virus nanoparticles. Nature Nanotech 4, 259–264 (2009). https://doi.org/10.1038/nnano.2009.38
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2009.38
This article is cited by
-
Engineering the gut microbiome
Nature Reviews Bioengineering (2023)
-
Development of low noise FET biosensor using graphene and ZnO nanostructures on cost effective substrates for biomolecule detection with enhanced performance
CSI Transactions on ICT (2020)
-
Recent advances in protein-based nanoparticles
Korean Journal of Chemical Engineering (2018)
-
Dual Electrophoresis Detection System for Rapid and Sensitive Immunoassays with Nanoparticle Signal Amplification
Scientific Reports (2017)
-
Synthesis, microstructure, and physical properties of metallic barcode nanowires
Metals and Materials International (2017)