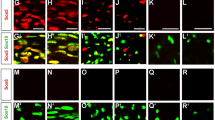
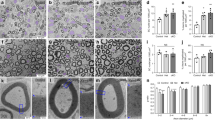
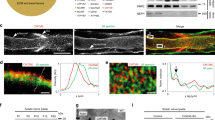
Abstract
Peripheral nerve development results from multiple cellular interactions between axons, Schwann cells and the surrounding mesenchymal tissue. The delayed axonal sorting and hypomyelination throughout the peripheral nervous system of claw paw (clp) mutant mice suggest that the clp gene product is critical for these interactions. Here we identify the clp mutation as a 225-bp insertion in the Lgi4 gene. Lgi4 encodes a secreted and glycosylated leucine-rich repeat protein and is expressed in Schwann cells. The clp mutation affects Lgi4 mRNA splicing, resulting in a mutant protein that is retained in the cell. Additionally, siRNA-mediated downregulation of Lgi4 in wild-type neuron–Schwann cell cocultures inhibits myelination, whereas exogenous Lgi4 restores myelination in clp/clp cultures. Thus, the abnormalities observed in clp mice are attributable to the loss of Lgi4 function, and they identify Lgi4 as a new component of Schwann cell signaling pathway(s) that controls axon segregation and myelin formation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Jessen, K.R. & Mirsky, R. Schwann Cell Development. in Myelin Biology and Disorders Vol. 1 (ed. Lazzarini, R.A.) 329–370 (Elsevier Academic, San Diego, 2004).
Lobsiger, C.S., Taylor, V. & Suter, U. The early life of a Schwann cell. Biol. Chem. 383, 245–253 (2002).
Britsch, S. et al. The ErbB2 and ErbB3 receptors and their ligand, neuregulin-1, are essential for development of the sympathetic nervous system. Genes Dev. 12, 1825–1836 (1998).
Garratt, A.N., Britsch, S. & Birchmeier, C. Neuregulin, a factor with many functions in the life of a Schwann cell. Bioessays 22, 987–996 (2000).
Michailov, G.V. et al. Axonal neuregulin-1 regulates myelin sheath thickness. Science 304, 700–703 (2004).
Taveggia, C. et al. Neuregulin-1 type III determines the ensheathment fate of axons. Neuron 47, 681–694 (2005).
Cosgaya, J.M., Chan, J.R. & Shooter, E.M. The neurotrophin receptor p75NTR as a positive modulator of myelination. Science 298, 1245–1248 (2002).
Chan, J.R. et al. NGF controls axonal receptivity to myelination by Schwann cells or oligodendrocytes. Neuron 43, 183–191 (2004).
Fields, R.D. & Stevens-Graham, B. New insights into neuron-glia communication. Science 298, 556–562 (2002).
Atanasoski, S. et al. The protooncogene Ski controls Schwann cell proliferation and myelination. Neuron 43, 499–511 (2004).
Ogata, T. et al. Opposing extracellular signal-regulated kinase and Akt pathways control Schwann cell myelination. J. Neurosci. 24, 6724–6732 (2004).
Wegner, M. Transcriptional control in myelinating glia: the basic recipe. Glia 29, 118–123 (2000).
Topilko, P. & Meijer, D. Transcription factors that control Schwann cell development and myelination. in Glial Cell Development (eds. Jessen, K.R. & Richardson, W.D.) 223–244 (Oxford Univ. Press, Oxford, 2001).
Ghislain, J. et al. Characterisation of cis-acting sequences reveals a biphasic, axon-dependent regulation of Krox20 during Schwann cell development. Development 129, 155–166 (2002).
Jaegle, M. et al. The POU proteins Brn-2 and Oct-6 share important functions in Schwann cell development. Genes Dev. 17, 1380–1391 (2003).
Le, N. et al. Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of Schwann cell differentiation and myelination. Proc. Natl. Acad. Sci. USA 102, 2596–2601 (2005).
Nagarajan, R. et al. EGR2 mutations in inherited neuropathies dominant-negatively inhibit myelin gene expression. Neuron 30, 355–368 (2001).
Britsch, S. et al. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 15, 66–78 (2001).
Henry, E.W., Eicher, E.M. & Sidman, R.L. The mouse mutation claw paw: forelimb deformity and delayed myelination throughout the peripheral nervous system. J. Hered. 82, 287–294 (1991).
Koszowski, A.G., Owens, G.C. & Levinson, S.R. The effect of the mouse mutation claw paw on myelination and nodal frequency in sciatic nerves. J. Neurosci. 18, 5859–5868 (1998).
Darbas, A. et al. Cell autonomy of the mouse claw paw mutation. Dev. Biol. 272, 470–482 (2004).
Bermingham, J.R., Jr. et al. Identification of genes that are downregulated in the absence of the POU domain transcription factor pou3f1 (Oct-6, Tst-1, SCIP) in sciatic nerve. J. Neurosci. 22, 10217–10231 (2002).
Gu, W. et al. The LGI1 gene involved in lateral temporal lobe epilepsy belongs to a new subfamily of leucine-rich repeat proteins. FEBS Lett. 519, 71–76 (2002).
Maro, G.S. et al. Neural crest boundary cap cells constitute a source of neuronal and glial cells of the PNS. Nat. Neurosci. 7, 930–938 (2004).
Staub, E. et al. The novel EPTP repeat defines a superfamily of proteins implicated in epileptic disorders. Trends Biochem. Sci. 27, 441–444 (2002).
Scheel, H., Tomiuk, S. & Hofmann, K. A common protein interaction domain links two recently identified epilepsy genes. Hum. Mol. Genet. 11, 1757–1762 (2002).
Buchanan, S.G. & Gay, N.J. Structural and functional diversity in the leucine-rich repeat family of proteins. Prog. Biophys. Mol. Biol. 65, 1–44 (1996).
Kajava, A.V. Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 277, 519–527 (1998).
Wong, K., Park, H.T., Wu, J.Y. & Rao, Y. Slit proteins: molecular guidance cues for cells ranging from neurons to leukocytes. Curr. Opin. Genet. Dev. 12, 583–591 (2002).
Howitt, J.A., Clout, N.J. & Hohenester, E. Binding site for Robo receptors revealed by dissection of the leucine-rich repeat region of Slit. EMBO J. 23, 4406–4412 (2004).
Gherardi, E., Love, C.A., Esnouf, R.M. & Jones, E.Y. The sema domain. Curr. Opin. Struct. Biol. 14, 669–678 (2004).
Skradski, S.L. et al. A novel gene causing a mendelian audiogenic mouse epilepsy. Neuron 31, 537–544 (2001).
Nakayama, J. et al. A nonsense mutation of the MASS1 gene in a family with febrile and afebrile seizures. Ann. Neurol. 52, 654–657 (2002).
Kalachikov, S. et al. Mutations in LGI1 cause autosomal-dominant partial epilepsy with auditory features. Nat. Genet. 30, 335–341 (2002).
Berkovic, S.F. et al. LGI1 mutations in temporal lobe epilepsies. Neurology 62, 1115–1119 (2004).
Ottman, R. et al. LGI1 mutations in autosomal dominant partial epilepsy with auditory features. Neurology 62, 1120–1126 (2004).
Pisano, T. et al. Abnormal phonologic processing in familial lateral temporal lobe epilepsy due to a new LGI1 mutation. Epilepsia 46, 118–123 (2005).
Gu, W., Sander, T., Becker, T. & Steinlein, O.K. Genotypic association of exonic LGI4 polymorphisms and childhood absence epilepsy. Neurogenetics 5, 41–44 (2004).
Ellgaard, L. & Helenius, A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4, 181–191 (2003).
Bonnon, C., Goutebroze, L., Denisenko-Nehrbass, N., Girault, J.A. & Faivre-Sarrailh, C. The paranodal complex of F3/contactin and caspr/paranodin traffics to the cell surface via a non-conventional pathway. J. Biol. Chem. 278, 48339–48347 (2003).
Senechal, K.R., Thaller, C. & Noebels, J.L. ADPEAF mutations reduce levels of secreted LGI1, a putative tumor suppressor protein linked to epilepsy. Hum. Mol. Genet. 14, 1613–1620 (2005).
Fernandez-Valle, C., Gorman, D., Gomez, A.M. & Bunge, M.B. Actin plays a role in both changes in cell shape and gene-expression associated with Schwann cell myelination. J. Neurosci. 17, 241–250 (1997).
Wolpowitz, D. et al. Cysteine-rich domain isoforms of the neuregulin-1 gene are required for maintenance of peripheral synapses. Neuron 25, 79–91 (2000).
Runkel, F., Michels, M. & Franz, T. Fxyd3 and Lgi4 expression in the adult mouse: a case of endogenous antisense expression. Mamm. Genome 14, 665–672 (2003).
McKay, B.E. & Turner, R.W. Physiological and morphological development of the rat cerebellar Purkinje cell. J. Physiol. (Lond.) 567, 829–850 (2005).
Bendtsen, J.D., Nielsen, H., von Heijne, G. & Brunak, S. Improved prediction of signal peptides: signalP 3.0. J. Mol. Biol. 340, 783–795 (2004).
Mayor, C. et al. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046–1047 (2000).
Archelos, J.J. et al. Production and characterization of monoclonal antibodies to the extracellular domain of P0. J. Neurosci. Res. 35, 46–53 (1993).
Zhuang, Y.A., Goldstein, A.M. & Weiner, A.M. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc. Natl. Acad. Sci. USA 86, 2752–2756 (1989).
Hart, G.W., Brew, K., Grant, G.A., Bradshaw, R.A. & Lennarz, W.J. Primary structural requirements for the enzymatic formation of the N-glycosidic bond in glycoproteins. Studies with natural and synthetic peptides. J. Biol. Chem. 254, 9747–9753 (1979).
Acknowledgements
We thank D. Krajacich for technical assistance, as well as the undergraduate summer students who worked on this project at the McLaughlin Research Institute: C. Schedl, M. Chen, S. Brauer, J. Anspach-Hanson and B. Cook. We also thank C. Ebeling for assistance with LI-COR analysis; S.S. Scherer for providing the PLP plasmid; N. Jenkins, N. Copeland, J. Chan and T. Watkins for protocols and advice; and E. Dzierzak for critical reading of the manuscript. This work was funded by the National Institute of Neurological Diseases and Stroke (NINDS) grant NS40751 and Muscular Dystrophy Association grant 3476 to J.R.B. Jr.; NINDS grants NS049087 and NS40745 to J.M.; and by grants from the Nederlandse Organisatie van Wetenschappelijk Onderzoek (NWO; ZonMW 903-42-195 and 901-01-205) to D.M.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Results of the C57BL/6J-clp/+ X BALB/c backcross. (PDF 643 kb)
Supplementary Fig. 2
Fine mapping of the clp candidate region. (PDF 688 kb)
Supplementary Fig. 3
BAC transgene complementation of Lgi4clp. (PDF 791 kb)
Supplementary Fig. 4
Mechanism of Lgi4clp exon exclusion. (PDF 703 kb)
Rights and permissions
About this article
Cite this article
Bermingham, J., Shearin, H., Pennington, J. et al. The claw paw mutation reveals a role for Lgi4 in peripheral nerve development. Nat Neurosci 9, 76–84 (2006). https://doi.org/10.1038/nn1598
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn1598
This article is cited by
-
Myelin Biology
Neurotherapeutics (2021)
-
Peripheral Nerve Development and the Pathogenesis of Peripheral Neuropathy: the Sorting Point
Neurotherapeutics (2021)
-
Spatial mapping of juxtacrine axo-glial interactions identifies novel molecules in peripheral myelination
Nature Communications (2015)
-
Transcriptome analysis of amoeboid and ramified microglia isolated from the corpus callosum of rat brain
BMC Neuroscience (2012)
-
The Cyclin-Dependent Kinase Inhibitor p27Kip1 is a Positive Regulator of Schwann Cell Differentiation In Vitro
Journal of Molecular Neuroscience (2011)