Abstract
Despite the variety of physiological and target-related functions, little is known regarding the cellular complexity in the sympathetic ganglion. We explored the heterogeneity of mouse stellate and thoracic ganglia and found an unexpected variety of cell types. We identified specialized populations of nipple- and pilo-erector muscle neurons. These neurons extended axonal projections and were born among other neurons during embryogenesis, but remained unspecialized until target organogenesis occurred postnatally. Target innervation and cell-type specification was coordinated by an intricate acquisition of unique combinations of growth factor receptors and the initiation of expression of concomitant ligands by the nascent erector muscles. Overall, our results provide compelling evidence for a highly sophisticated organization of the sympathetic nervous system into discrete outflow channels that project to well-defined target tissues and offer mechanistic insight into how diversity and connectivity are established during development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Change history
05 September 2016
In the version of this article initially published online, the symbols were reversed in the key for Figure 2l. The error has been corrected for the print, PDF and HTML versions of this article.
References
Cannon, W.B. Organization for physiological homeostasis. Physiol. Rev. 9, 399–431 (1929).
Goldstein, D.S. Differential responses of components of the autonomic nervous system. in Handbook of Clinical Neurology (eds. J.C. Masdeu & R.G. Gonzalez) (Elsevier, 2013).
Langley, J.N. The Autonomic Nervous System (W. Heffer and Sons, 1921).
Selye, H. Stress Without Distress (New American Library, 1974).
Robertson, D. et al. Comparative assessment of stimuli that release neuronal and adrenomedullary catecholamines in man. Circulation 59, 637–643 (1979).
Pacak, K. et al. Heterogeneous neurochemical responses to different stressors: a test of Selye's doctrine of nonspecificity. Am. J. Physiol. 275, R1247–R1255 (1998).
Jänig, W. Integrative Action of the Autonomic Nervous System (Cambridge University Press, 2006).
Clapham, J.C. Central control of thermogenesis. Neuropharmacology 63, 111–123 (2012).
Ootsuka, Y. & Blessing, W.W. Inhibition of medullary raphé/parapyramidal neurons prevents cutaneous vasoconstriction elicited by alerting stimuli and by cold exposure in conscious rabbits. Brain Res. 1051, 189–193 (2005).
Benedek, M. & Kaernbach, C. Physiological correlates and emotional specificity of human piloerection. Biol. Psychol. 86, 320–329 (2011).
Birkenfeld, A. & Kase, N.G. Functional anatomy and physiology of the female breast. Obstet. Gynecol. Clin. North Am. 21, 433–444 (1994).
Uvnäs-Moberg, K. & Eriksson, M. Breastfeeding: physiological, endocrine and behavioural adaptations caused by oxytocin and local neurogenic activity in the nipple and mammary gland. Acta Paediatr. 85, 525–530 (1996).
Vorherr, H. The Breast: Morphology, Physiology, and Lactation (Academic Press, 1974).
Lindquist, C., Fedorcsak, I. & Steig, P.E. Electrophysiological aid in high thoracic sympathectomy for palmar hyperhidrosis. Neurosurgery 24, 449–452 (1989).
Gibbins, I. Functional organization of autonomic neural pathways. Organogenesis 9, 169–175 (2013).
Ernsberger, U. The development of postganglionic sympathetic neurons: coordinating neuronal differentiation and diversification. Auton. Neurosci. 94, 1–13 (2001).
Elfvin, L.G., Lindh, B. & Hökfelt, T. The chemical neuroanatomy of sympathetic ganglia. Annu. Rev. Neurosci. 16, 471–507 (1993).
Gibbins, I.L. Vasomotor, pilomotor and secretomotor neurons distinguished by size and neuropeptide content in superior cervical ganglia of mice. J. Auton. Nerv. Syst. 34, 171–183 (1991).
Zeisel, A. et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015).
van der Maaten, L. & Hinton, G.E. Visualizing high-dimensional data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Tanabe, Y. & Jessell, T.M. Diversity and pattern in the developing spinal cord. Science 274, 1115–1123 (1996).
Briscoe, J. & Ericson, J. Specification of neuronal fates in the ventral neural tube. Curr. Opin. Neurobiol. 11, 43–49 (2001).
Burau, K. et al. c-ret regulates cholinergic properties in mouse sympathetic neurons: evidence from mutant mice. Eur. J. Neurosci. 20, 353–362 (2004).
Furlan, A., Lübke, M., Adameyko, I., Lallemend, F. & Ernfors, P. The transcription factor Hmx1 and growth factor receptor activities control sympathetic neurons diversification. EMBO J. 32, 1613–1625 (2013).
Gonsalvez, D.G. et al. Proliferation and cell cycle dynamics in the developing stellate ganglion. J. Neurosci. 33, 5969–5979 (2013).
Holzmann, J., Hennchen, M. & Rohrer, H. Prox1 identifies proliferating neuroblasts and nascent neurons during neurogenesis in sympathetic ganglia. Dev. Neurobiol. 75, 1352–1367 (2015).
Luo, W., Enomoto, H., Rice, F.L., Milbrandt, J. & Ginty, D.D. Molecular identification of rapidly adapting mechanoreceptors and their developmental dependence on ret signaling. Neuron 64, 841–856 (2009).
Airaksinen, M.S. & Saarma, M. The GDNF family: signaling, biological functions and therapeutic value. Nat. Rev. Neurosci. 3, 383–394 (2002).
Clarke, J. et al. Interaction of neuropeptide Y and the sympathetic nervous system in vascular control in man. Circulation 83, 774–777 (1991).
Lundberg, J.M. et al. Neuropeptide Y (NPY)-like immunoreactivity in peripheral noradrenergic neurons and effects of NPY on sympathetic function. Acta Physiol. Scand. 116, 477–480 (1982).
Damon, D.H., Teriele, J.A. & Marko, S.B. Vascular-derived artemin: a determinant of vascular sympathetic innervation? Am. J. Physiol. Heart Circ. Physiol. 293, H266–H273 (2007).
Honma, Y. et al. Artemin is a vascular-derived neurotropic factor for developing sympathetic neurons. Neuron 35, 267–282 (2002).
Wolf, E., Black, I.B. & DiCicco-Bloom, E. Mitotic sympathetic neuroblasts initiate axonal pathfinding in vivo. J. Neurobiol. 40, 366–374 (1999).
Schuchardt, A., D'Agati, V., Larsson-Blomberg, L., Costantini, F. & Pachnis, V. Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature 367, 380–383 (1994).
Baudet, C. et al. Retrograde signaling onto Ret during motor nerve terminal maturation. J. Neurosci. 28, 963–975 (2008).
Francis, N.J. & Landis, S.C. Cellular and molecular determinants of sympathetic neuron development. Annu. Rev. Neurosci. 22, 541–566 (1999).
Schotzinger, R., Yin, X. & Landis, S. Target determination of neurotransmitter phenotype in sympathetic neurons. J. Neurobiol. 25, 620–639 (1994).
Gibbins, I.L. & Morris, J.L. Sympathetic noradrenergic neurons containing dynorphin but not neuropeptide Y innervate small cutaneous blood vessels of guinea-pigs. J. Auton. Nerv. Syst. 29, 137–149 (1990).
Schneider, C., Wicht, H., Enderich, J., Wegner, M. & Rohrer, H. Bone morphogenetic proteins are required in vivo for the generation of sympathetic neurons. Neuron 24, 861–870 (1999).
Huang, T. et al. Tlx3 controls cholinergic transmitter and peptide phenotypes in a subset of prenatal sympathetic neurons. J. Neurosci. 33, 10667–10675 (2013).
Bonanomi, D. et al. Ret is a multifunctional coreceptor that integrates diffusible- and contact-axon guidance signals. Cell 148, 568–582 (2012).
Charoy, C. et al. gdnf activates midline repulsion by Semaphorin3B via NCAM during commissural axon guidance. Neuron 75, 1051–1066 (2012).
Uesaka, T., Nagashimada, M., Yonemura, S. & Enomoto, H. Diminished Ret expression compromises neuronal survival in the colon and causes intestinal aganglionosis in mice. J. Clin. Invest. 118, 1890–1898 (2008).
Parlato, R., Otto, C., Begus, Y., Stotz, S. & Schütz, G. Specific ablation of the transcription factor CREB in sympathetic neurons surprisingly protects against developmentally regulated apoptosis. Development 134, 1663–1670 (2007).
Hippenmeyer, S. et al. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 3, e159 (2005).
Harvey, N.L. et al. Lymphatic vascular defects promoted by Prox1 haploinsufficiency cause adult-onset obesity. Nat. Genet. 37, 1072–1081 (2005).
Rossi, J. et al. Retarded growth and deficits in the enteric and parasympathetic nervous system in mice lacking GFR alpha2, a functional neurturin receptor. Neuron 22, 243–252 (1999).
Danielian, P.S., Muccino, D., Rowitch, D.H., Michael, S.K. & McMahon, A.P. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 8, 1323–1326 (1998).
Drucker, H., Burges, C.J.C., Kaufman, L., Smola, A. & Vapnik, V. Support vector regression machines. Adv. Neural Inf. Process. Syst. 6, 155–161 (1997).
Pedregosa, F.V.G. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Jones, E., Oliphant, T. & Peterson, P. SciPy: Open source scientific tools for Python. http://www.scipy.org/ (2001).
Mi, H., Muruganujan, A. & Thomas, P.D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–D386 (2013).
Pirici, D. et al. Antibody elution method for multiple immunohistochemistry on primary antibodies raised in the same species and of the same subtype. J. Histochem. Cytochem. 57, 567–575 (2009).
Meijering, E. et al. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A 58, 167–176 (2004).
Acknowledgements
We thank S. Arber (Friedrich Miescher Institute for Biomedical Research) for kindly providing the TaumGFP mouse line, and J.-F. Brunet (Institut de Biologie de l'École Normale Supérieure), T. Müller (The Max Delbrück Center for Molecular Medicine) and T. Taija Mäkinen (Uppsala University) for generously providing PHOX2B, TLX3 and PROX1 antibodies, respectively. We also thank S. Lücke and J. Sontheimer for technical support, and J. Hjerling-Leffler (Karolinska Institutet) for reagents. This work was supported by the Swedish Medical Research Council, Knut and Alice Wallenbergs Foundation (Wallenberg Scholar and for CLICK imaging facility), SFO grant (DBRM), Swedish Cancer Foundation, Söderbergs Foundation and Karolinska Institutet (P.E.), Knut and Alice Wallenberg's Foundation (2015.0041), Swedish Research Council (STARGET), Swedish foundation for strategic research (RIF14-0057) (to S.L.).
Author information
Authors and Affiliations
Contributions
A.F. and P.E. designed the study. P.E. supervised the study and S.L. supervised the single-cell RNA-seq experiments. A.F. performed most of the experiments, analyzed the data and prepared the figures. A.F., M.H., G.L.M., H.H. and D.U. dissected, dissociated captured SG single cells and prepared cDNA libraries. G.L.M. performed single-cell analysis and prepared these figures. M.L. bred TrkACre, RetCFP and Retfl mouse lines and processed the tissues. M.L. and H.A. performed fate-tracing experiments. J.K. and M.S.A. provided Gfra2KO tissue. G.O. provided Prox1floxed mice. A.F. and P.E. wrote the paper, with input from all of the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Single-cell RNA sequencing reveals unique sets of neuropeptides, transcription factors and receptors in specific types of sympathetic neurons
(a) Dissection of paravertebral SGs was carried out using Wnt1-Cre;R26RTOMATO mice. All organs, except the dorsal aorta (asterisks) were removed, leaving an empty rib cage (ventral view). Thoracic SGs are essentially invisible to the naked eye (left panel, brightfield) but they become visible when fluorescent light is used (right panel). Arrowheads point at the site of the SGs. (b-d) Isolated single sympathetic neurons. (b) Representative images of group-specific sympathetic neurons (in red) captured in the C1 chip. Histograms showing soma area (c) and molecule counting distributions (d) of individual cells (black bars) and average within each group of neurons (red line). (e-g) The binarized expression of transcription factors, receptors and neuropeptides. Rectangles are drawn next to single or multiple cell types to represent binary patterns of expression. For each pattern, the names of genes expressed at posterior probability > 99% levels above baseline are indicated. (h) Extended heat map showing average expression levels of genes shown in Fig. 1, e-g. Colormap: blue, the gene is expressed above baseline (> 99% posterior probability); grey, the gene is not expressed above baseline. Scale bar: 1mm (a).
Supplementary Figure 2 In vivo confirmation of the existence of diverse sympathetic types predicted by single-cell RNA sequencing.
(a) Schematic representation of neuronal types with markers used for validation. Genes between brackets were predicted from RNAseq but not validated. (b) Sympathetic neuronal types verified in vivo by a combinatorial immunohistochemical approach. NA1-5 neurons all express TH and TRKA; ACh1-2 express VACHT and GFRα2; Glut neurons express VGLUT2. NA1 express RARRES1; NA2 and NA3 express NPY; NA2 but not NA3 are RET+ and Htr3aEGFP+, therefore NA2 are NPY+ RET+Htr3aEGFP+ and NA3 are NPY+RET-Htr3aEGFP-. NA2 also express GFRα3. NA4-5 was separated from NA1-3 based on their expression of GFRα2. NA4 and NA5 neurons were separated based on expression of ENC1. Hence, NA4 are GFRα2+ENC1+, whereas NA5 are GFRα2+ENC1-. ACh1 were separated from ACh2 neurons based on expression of SST. Ret expression was present at low levels (Retlow) in NA2 and NA4-5 but not in NA3. NA1 neurons were mostly RET- (see also Supplementary Fig. 1d). ACh1 and ACh2 express Ret at high levels (Rethigh). (c) Violin plots showing the mRNA expression of the selection of genes used to validate the existence of predicted neuronal groups. (d-t) Extended validation of neuronal types. (d) Triple immunohistochemistry for NPY, TOMATO (recapitulating TrkA expression) and RARRES1 (upper panel) and RET (lower panel) on adult SGs from TrkACre/+;R26RTOM/+ animals. Sections were first incubated and imaged with the first set of primary antibodies (rabbit anti-NPY, chicken anti-TOMATO and goat anti-RARRES1), then eluted (Glycine+SDS pH2, see Methods), then re-incubated with the second set of primary antibodies (rabbit anti-NPY, chicken anti-TOMATO and goat anti-RET). Inset and arrowheads show that NA1 RARRES1+NPY- neurons are generally negative for RET expression. NA1 neurons occasionally expressed NPY (20% of all RARRES1+ cells) and RET (30% of all RARRES1+ cells) but never together. (e) Triple immunohistochemistry for FST, NPY and GFP (recapitulating Htr3a expression) on P35 SGs from Htr3a-EGFP mice. Upper panel: FST+NPY- NA1 neurons express Htr3a (inset) whereas NA3 neurons are FST+NPY+Htr3aEGFP- (arrowheads); lower panel: NA2 neurons are FST+NPY+Htr3aEGFP+(inset). (f) Triple immunohistochemistry for GFRα3, GFP (recapitulating Ret expression) and NPY on adult SGs from RetCFP/+ mice shows NA2 cells are GFRα3+Retlow+NPY+ (inset). (g) Triple immunohistochemistry for NPY, RET and GFP (recapitulating Htr3a expression) on P35 SGs from Htr3a-EGFP mice. NA2 cells (inset NA2) are NPY+RET+Htr3aEGFP+, whereas NA3 cells (inset NA3) are NPY+ only. (h) Triple immunohistochemistry for GAL, NPY and GFP (recapitulating Htr3a expression) on on P35 SGs from Htr3a-EGFP mice. NA2 (inset) and NA3 cells (arrowheads) express GAL. (i) Triple immunohistochemistry for ESR1, NPY and GFP (recapitulating Htr3a expression) on P35 SGs from Htr3a-EGFP mice shows ESR1 is expressed in NA2 cells (inset). (j) Triple immunohistochemistry for CDH8, NPY and FST (both in green) and TH on SGs from adult wild-type mice. NA1-3 neurons do not express CDH8, which is expressed uniquely by NA4-5 groups (inset). (k) Triple immunohistochemistry for CDH8, GFP (recapitulating Ret expression) and TRKA on P11 SGs from RetCFP mice shows NA4-5 CDH8+ cells express Retlow (inset). (l) Triple immunohistochemistry for TYRO3, GFP (recapitulating Ret expression) and TH on adult SGs from RetCFP mice shows NA4-5 Retlow cells express TYRO3 (inset). TYRO3 is also expressed by ACh1-2 (TH-Rethigh neurons, arrowheads). (m) Triple immunohistochemistry for GFRα2, GFP (recapitulating Ret expression) and NPY on adult SGs from RetCFP mice shows NA4-5 Retlow cells express GFRA2. GFRA2 is expressed also by ACh1-2 (NPY- Rethigh neurons, arrowheads). (n) Triple immunohistochemistry for ENC1, TOMATO (recapitulating TrkA expression) and GFRΑ2 on adult SGs from TrkACre/+;R26RTOM/+ animals shows that NA5 GFRα2+ ENC1-negative cells (arrowheads) could be distinguished from ENC1-expressing NA4 cells (inset). (o) Triple immunohistochemistry for CNTN6, CDH8, and TH on P11 SGs from wild-type animals shows that NA5 GFRα2+ CNTN6-negative cells (arrowheads) cells can be distinguished from CNTN6-expressing NA4 cells (inset). (p) Triple immunohistochemistry for ENC1, GFP (recapitulating Ret expression) and TH on adult SGs from RetCFP/+ mice confirms that both NA4 ENC1+ (inset) and NA5 ENC1- (arrowheads) cells express Retlow. (q) Triple immunohistochemistry for SST, VACHT (Slc18a3) and DAPI on adult SGs from Wnt1-Cre;R26RTOM/+ animals shows that cholinergic neurons are divided into VACHT+SST- (ACh1 group) and VACHT+SST+ (ACh2 group) neurons (inset). (r) Triple immunohistochemistry for VACHT, GFP (recapitulating Ret expression) and VIP on adult SGs from RetCFP mice shows ACh1-2 VACHT+ neurons express Rethigh and VIP (inset). (s) Triple immunohistochemistry for VACHT, TH and ISL1 on P8 SGs from wild-type animals shows ISL1+ neurons that are associated to the sympathetic ganglion but do not express either TH or VACHT (arrowheads), thus they do not belong to NA1-5 or ACh1-2. (t) Double immunohistochemistry for VGLUT2 (Slc17a6) and TH on P8 SGs from wild-type animals shows that TH- neurons (arrowheads) express VGLUT2 and are therefore glutamatergic. The staining was carried out on a section consecutive of the one shown in (r). (u) Proportion of identified sympathetic cell types in the superior cervical ganglion (SCG), stellate ganglion and thoracic ganglia 1-4 and 5-12. In the SCG, only 55.8% of all neurons could be identified to belong to either NA1-5 or ACh1-2 neuronal types. Thus, about half of these neurons belong to classes of neurons yet to be discovered. (v) Soma size of sympathetic types. NA2, NA4 and NA5 show a significantly larger soma size compared to NA1, NA3, ACh1 and ACh2 (F(6, 14) = 30.94, P < 0.0001, one-way analysis of variance, n=3. Post-test: BonferroniÕs multiple comparison test. All data are presented as mean ± s.e.m.). (w) Double immunohistochemistry for NPY and RET on adult stellate sections from DbhCre;R26Tomato mice. Note NA2 neurons clustered together within the ganglion (dashed line). (b), (d-t) and (w) are representative images from experiments carried out on at least two animals. Scale bars: 5 μm (b), 20 μm (d-t), 50 μm (w).
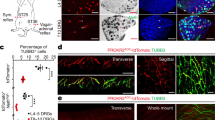
Supplementary Figure 3 Erector muscle organogenesis commences shortly after birth and receives sympathetic innervation a few days thereafter.
(a) Double immunohistochemistry for ASMACy3 and K5 (an epidermal keratin detected by anti-keratin 5) on P1 and P3 thoracic skin sections from wild-type mice. NEM organogenesis takes place between P1 and P3 (arrowheads). ASMACy3+ tissues at P1 are blood vessels. (b) Triple immunohistochemistry for ASMACy3, TH and K5 on P5, P7 and P11 NEM sections from wild-type mice. Note that TH+ axons innervating the NEM increase in density from P5 to P11 (arrowheads). (c) Triple immunohistochemistry for NPY, TH and RET on NEM sections from adult wild-type mice. Note TH+NPY+RET+ sympathetic axons (insets). Insets are single optical sections selected from a Z-stack of the region of interest, acquired at 2 μm intervals. (d) Triple immunohistochemistry for TH, GFP and PGP9.5 (protein gene product 9.5, encoded by ubiquitin-C-terminal hydrolase 1, to detect all nerve fibers) on adult NEM sections from RetCreERT2;R26RmGFP shows TH and RetmGFP expression in different location of the PGP9.5+ fiber. Presence of both markers suggests a RET+TH+ origin. (e) Innervation at the base of NEM (ASMACy3+) is initiated at P6 from TH+RetCFP+ fibers (inset). (f) Double immunohistochemistry for ASMACy3 and K5 on P1 and P3 dorsal skin sections from wild-type mice. PEM organogenesis takes place between P1 and P3 (arrowhead). (g-j) Triple immunohistochemistry for ASMACy3, TH and K5 or CD31 on P3, P5, P7 and P11 PEM sections from wild-type mice. Note TH+ sympathetic axons (arrows) in close association with CD31+ blood vessels (g-h, arrowheads), located in proximity of the PEM (asterisks), suggesting that PEM innervation is received from nerves associated with nearby blood vessels. Innervation of PEM is initiated at P7 (i, insets) and it is robust at P11 (j). (k) Triple immunohistochemistry for TH, NPY and ASMACy3 on P20 PEM sections from RetCFP mice. TH+ sympathetic axons (arrows) innervating the PEM do not show immunoreactivity for NPY, showing that PEM is not a target of NA2 or NA3 neurons. Insets are max intensity projections of a Z-stack of the region of interest, acquired at 2 μm intervals. (l) Innervation of PEM is initiated at P6; TH+RetCFP+ fibers are present on blood vessels (arrows, inset) and on the base of PEM (arrowheads, inset). (m) Sympathetic DbhTOM+ traced NEM fibers (in DbhCre/+;R26RTOM/+ mice) express general noradrenergic markers such as TRKA (inset) and NA1-NA3 markers, such as GFRα3 (arrowheads). (n) RetCFP+ (in RetCFP mice, P20) PEM fibers contain TRKA (inset). (o-s) FastBlue (FB) retrograde tracing of axons innervating NEM (o, p) and PEM (q-s). (o) NEM injection site. Double immunohistochemistry for ASMA and TH on NEM samples from Wnt1-Cre;R26RTOM/+ adult mice injected with FB (green signal). The nipple is outlined, arrowheads point at NEM. (p)Triple immunohistochemistry for NPY, RET and TOMATO on SG of traced Wnt1-Cre;R26RTOM/+adult mice. Traced cells express both NPY and RET, matching the molecular signature of NA2 neurons. (q) PEM injection site. Double immunohistochemistry for ASMACy3 and K5 on back skin samples from TrkACre;R26RTOM/+ adult mice injected with FB (green signal). Arrowheads point at PEMs. (r, s) Triple immunohistochemistry for RET, NPY and TOMATO (recapitulating TrkA expression, r) and for ENC1, GFRα2 and TOMATO (s) on SG from traced TrkACre/+;R26RTOM/+ adult mice. Note traced FB+TrkATOM+ neurons in (r) express RET (arrowheads) but are negative for NPY and that FB+TrkATOM+GFRα2+ neurons in (s) are negative for ENC1. (t, u) Quantification of FB+ traced cells following injection of NEM (t) or PEM (u). FB+ cells expressing NPY and RET (NPY+RET+) were classified as NA2, those expressing only NPY (NPY+RET-) as NA3 and those expressing only RET (NPY-RET+) as NA4-5 neurons. FB+ cells that were negative for both markers were labeled as ÒunassignedÓ. Neurons classified as NA2 were virtually absent when the tracer was injected in the dorsal skin PEMs (t) while when injected into the NEMs traced neurons represent more than one third of all traced cells (u). The skin is richly innervated by autonomic neurons of various types, including cutaneous blood vessels, nipple and hair follicle erector muscles. Thus, as shown in (o) and (q), injecting 0.25 ml FB results in some spread of the tracer, resulting in traced neurons also from nearby autonomic targets. Therefore, in (t), we conclude that the remaining 70% of FB+ neurons are traced from blood vessels (NA3 neurons) or from ventral hair follicle PEMs (NA4-5 neurons) of the tissue surrounding the nipple; in (u), the greatest part of FB+ cells are NA4-5 and the second most abundant group is represented by presumed blood vessel NA3 neurons. (a-m), (n), (o) and (q) are representative images from experiments carried out on at least two animals. Scale bars: 100 μm in (o) and (r); 20 μm in (a-c) and (e-l); 10 μm in (n), (p), (q), (s); 2 μm in (d), (m) and all insets.
Supplementary Figure 4 NA2 and NA4-5 neurons specialize postnatally; PROX1 labels actively proliferating neuroblasts in the SG.
(a) Double immunohistochemistry for RET and TRKA on P3, P5, P6, P7, P8 and P11 SGs from wild-type mice shows emergence of RET+TRKA+ neurons during postnatal development. Quantification of the proportion of TRKA+RET+ neurons in the postnatal SG. TRKA+RET+ but not TRKA-only+ neurons exhibit a selective soma area growth (b, P5 vs P6: t(4) = 17.06, P <0.0001; P6 vs P7: t(4) = 10.40, P = 0.0005; P7 vs P8: t(4) = 7.294, P = 0.0019; P8 vs P11: t(4) = 1.474, P = 0.2145; P11 vs Adult: t(4) = 1.452, P = 0.2202; unpaired t test, n=3) and soma size (c, P5 vs P6: t(4) = 6.384, P = 0.0031; P6 vs P7: t(4) = 6.446, P = 0.0030; P7 vs P8: t(4) = 6.361, P = 0.0031; P8 vs P11: t(4) = 2.166, P = 0.0962; P11 vs Adult: t(4) = 1.456, P = 0.2190; unpaired t test, n=3. All data are presented as mean ± s.e.m.). (d-g) EMNs markers are largely absent before birth. (d) Triple immunohistochemistry for FST, a marker of NA1-NA3 neurons, NPY and TH on SG sections from E18.5 and P11 WT mice. FST+ neurons are increased at P11, compared to E18.5. (e) Quantification of (d): (t(4) = 15.9, P < 0.0001, unpaired t test, n=3. Data are presented as mean ± s.e.m.). (f-g) Double immunohistochemistry for TYRO3, a marker of NA4-5 neurons (f) or CNTN6 (g), a marker of NA4 cells, and TH on SG sections from E18.5 and P11 WT mice. Expression of both markers is absent at E18.5 but present at P11. (h) Triple immunohistochemistry for PROX1, TH and VACHT on SG sections from E12.5 to P11 WT mice. The proportion of PROX1+TH+ neuroblasts decreases during development. (i) Quantification of (h). n = 2-3 per stage, data are presented as mean ± s.e.m.. (j, k) Triple immunohistochemistry for PROX1 and EdU and KI67 (j) or PH3 and ISL1 (k) on SG sections from P0 wild-type mice. Most or all PROX1+ cells were KI67+. PROX1+ cells incorporate EdU and contain PH3, which is consistent with proliferation. (l) Double immunohistochemistry for PROX1 and EdU on SG sections from E12.5, E15.5 and E18.5 wild‑type mice showing that PROX1+ cells incorporate EdU at all analyzed stages. (m) Quantification of (l). n = 2-3 per stage, data are presented as mean ± s.e.m.. (a), (f), (g), (j) (o) and (q) are representative images from experiments carried out on at least two animals. Scale bars: 10 μm in (a); 20 μm in (h), (d), (f), (g) and (j-l).
Supplementary Figure 5 EMNs are induced postnatally from TRKA+ sympathetic neurons.
(a) Illustration of cell fate-tracing strategies. Neonatal mice were injected either at P0 with EdU, or at P3 with Tamoxifen (TAM). The efficiency of recombination of RetCreERT2;R26RTOMATO and TrkACreERT2;R26RTOMATO mouse lines was examined at P5 and analyses were performed in animals sacrificed at P11. (b) Triple immunohistochemistry for NPY, EdU and RET on SG sections from P11 wild-type mice injected with EdU at P0. RET+NPY+ neurons (inset) were rarely EdU+ (arrowheads). (c-f) Recombination efficiency and validation of the RetCreERT2;R26RTOMATO mouse strain for tracing of cholinergic neurons. (c) Double immunohistochemistry for PROX1 and GFP on SG sections from P3 RetCFP mice. PROX1+ cells expressed low levels of RET (RetlowPROX1+ cells, arrow), when compared to cholinergic RethighPROX1- cells (arrowhead). (d) Double immunohistochemistry for TOMATO (recapitulating RetTOM) and cholinergic markers VIP or TLX3, on SG sections from P5 RetCreERT2;R26RTOMATO mice injected with Tamoxifen at P3, showing TOMATO+ cells expressing markers of the cholinergic lineage of sympathetic neurons. (e) Triple immunohistochemistry for TOMATO (recapitulating RetTOM), PROX1 and VACHT on SG sections from P5 RetCreERT2;R26RTOMATO mice, injected with Tamoxifen at P3. The vast majority of RetTOM+ cells at P5 were VACHT+ showing that this line can be used to trace cells of a cholinergic origin. In rare cases, recombination occurred in PROX1+VACHT- cells expressing low levels of Ret (insets). (f) Quantification of (e). n = 2, data are presented as mean ± s.e.m.. Percentages refer to the proportion of each cell type within the total of RetTOM+ neurons. (g) Generation of a TrkACreERT2 mouse strain. The black boxes represent the exons of the Ntrk1 (TrkA) gene. An internal ribosomal entry site (IRES) directly followed by CreERT2 was introduced after the stop codon of the TrkA gene. The neomycin selection cassette was eliminated by Flp recombination to generate the TrkAIRES‑CreERT2 mouse line (abbreviated TrkACreERT2 line). (h-j) Recombination specificity of TrkACreERT2;R26RTOMATO mice. (h) Double immunohistochemistry for TOMATO (recapitulating TrkATOM) and PROX1 or VACHT, VIP, TLX3, confirms that TrkATOM cells are neither dividing cells nor cholinergic neurons. (i) All TrkATOM cells at P5 are TRKA+ and, very rarely, RET+ (arrowheads). (j) Quantification of (i). n = 3 per stage, data are presented as mean ± s.e.m.. Percentages in (j) refer to the proportion of each cell type within the total number of TrkATOM+ neurons. (b-d) and (h) are representative images from experiments carried out on at least two animals. Scale bars: 10 μm (c), (d), (i), (j); 20 μm (b), (e), (f).
Supplementary Figure 6 NA2 and NA4-5 neurons display different pathways of development.
(a) Triple immunohistochemistry for GFRα3, GFP (recapitulating Ret expression) and TRKA on P3, P5 and P11 SG sections from RetCFP mice. At P3, GFRα3 is expressed in a subset of TRKA+ neurons, while RetCFP and TRKA expression are mutually exclusive. Starting from P5, RetCFP expression is upregulated in a subset of GFRα3+TRKA+ neurons. (b) Triple immunohistochemistry for GFRα2, GFP (recapitulating Ret expression) and TRKA on P3, P5 and P11 SG sections from RetCFP mice. Neither GFRα2 nor RetCFP is expressed in noradrenergic TRKA+ neurons at P3 but GFRα2+RetCFP+ neurons are present at P5 and they increase in numbers at P11. GFRα2+TRKA- neurons are cholinergic neurons. (c) Triple immunohistochemistry for GFRα3, GFP (recapitulating Ret expression) and NPY on P11 SG sections from RetCFP mice. NA2 neurons express RetCFP+, NPY and GFRα3 (inset). (d) Triple immunohistochemistry for GFRα2, NPY and TH on P11 SG sections from RetCFP mice. NA4-5 neurons express GFRα2 and TH but are negative for NPY (inset). (e-f) NPY expression during development. (e) Quantification of NPY+ over TH+ neurons shows that the number of NPY+ neurons does not change between P3 and P11. t(10) = 0.4284, P = 0.6774; unpaired t test, n=6. (f) The percentage of RetCFP+NPY+ neurons increases from P3 to P11. n = 3 per stage, data are presented as mean ± s.e.m.. (g) Illustration summarizing these results. (a-d) are representative images from experiments carried out on at least two animals. Scale bars: 20 μm for all images.
Supplementary Figure 7 The onset of ARTN and NRTN expression in the nascent erector muscles is paralleled by downregulation of ARTN in the blood vessels and coincides with target innervation by sympathetic nerves.
(a) Quadruple immunohistochemistry for ARTN, TH, ASMACy3 and CD31 on P3 and P11 NEM sections from wild type mice. TH+ sympathetic axons were observed associated with blood vessels (arrowheads) and at the base of the NEM at P3 and on the ARTN-expressing NEM at P11 (arrows). (b) Triple immunohistochemistry for NRTN, TH and ASMACy3 on P5 and P7 PEM sections from wild type mice. Note TH+ sympathetic axons (arrowheads) at the base of the PEM at P5 and on NRTN expressing PEM at P7 (arrow). (c) Quadruple immunohistochemistry for ASMACy3, ARTN, CD31 and DAPI on P5 NEM sections from wild-type mice. CD31+ASMACy3+ blood vessel cells (arrows) in close association with the NEM express lower levels of ARTN, when compared to ASMACy3+ NEM cells (arrowheads). (d) Quadruple immunohistochemistry for ASMACy3, ARTN, K5 and DAPI on adult NEM sections from wild type mice. ASMACy3+ blood vessel cells (dashed lines) in close association with the NEM express little or no ARTN. (a-d) are representative images from experiments carried out on at least two animals. Scale bars: 10 μm in all images.
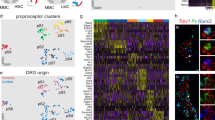
Supplementary Figure 8 Premature cell cycle exit and differentiation of SG neurons to EMNs in PROX1-deficient mice.
(a) Double immunohistochemistry for PROX1 and PHOX2B on SG sections from E14.5 control and Wnt1-Cre;Prox1f/f (Prox1cKO/cKO) mice. PROX1 is efficiently deleted in all SG neurons of Prox1cKO/cKO embryos, as shown by complete loss of PROX1 immunoreactivity in the mutant. (b) Double immunohistochemistry for PHOX2B and active CASPASE-3 (CASP3) on SG sections from E13.5 control and Prox1cKO/cKO mice. No apoptosis was detected in the SG, based on active CASPASE-3 staining, while some cells were positive in the dorsal root ganglion, which served as positive control tissue (inset). (c) Triple immunohistochemistry for KI67, EdU and PHOX2B on SG sections from E13.5 control and Prox1cKO/cKO mice shows a decreased proportion of proliferating cells in mutant mice. (d) Quantification of number of neurons per ganglion in E14.5 control and Prox1cKO/cKO embryos. (t(112) = 6.426, P < 0.0001, unpaired t test, 1 ganglion = 1 dot; 19 ganglia per animal were plotted, n=3). (e) EdU incorporation in control and Prox1cKO/cKO embryos. E13.5: t(3) = 6.587, P = 0.0071, unpaired t test, n=2 for control and n=3 for Prox1cKO/cKO; E14.5: t(4) = 6.659, P = 0.0026, unpaired t test, n=3; E18.5: t(4) = 5.084, P = 0.0071, unpaired t test, n=3. (f) Proportion of KI67+ neuroblasts in control and Prox1cKO/cKO embryos at E13.5. (t(3) = 5.845, P = 0.0100, unpaired t test, n=2 for control and n=3 for Prox1cKO/cKO). (g) Rate of cell division in control and Prox1cKO/cKO embryos at E13.5 (t(3) = 1.826, P = 0.1653, unpaired t test, n=2 for control and n=3 for Prox1cKO/cKO). All data are presented as mean ± s.e.m.. (h) Double immunohistochemistry for RET and TRKA on SG sections from E14.5 and E18.5 control and Prox1cKO/cKO reveals a massive increase of RET+TRKA+ neurons at both stages. (i) Triple immunohistochemistry for RET, NPY and TH on SG sections from E18.5 control and Prox1cKO/cKO mice. In control, NPY+TH+ neurons are RET-negative while in mutant mice, RET is upregulated in NPY+TH+ (e.g. NEM) neurons (arrowheads and insets). (j-m) Quantifications of (h) and (i). (j) Proportion of TRKA+ neurons at E14.5 and E18.5 in control and Prox1cKO/cKO mice. E14.5: t(9) = 4.552, P = 0.0014, unpaired t test, n=6 for control and n=5 for Prox1cKO/cKO; E18.5: t(4) = 13.12, P = 0.0002, unpaired t test, n=3. (k) Proportion of RET+ neurons at E14.5 and E18.5 in control and Prox1cKO/cKO mice. E14.5: t(9) = 2.366, P = 0.0422, unpaired t test, n=6 for control and n=5 for Prox1cKO/cKO; E18.5: t(4) = 4.525, P = 0.0106, unpaired t test, n=3. (l), TRKA+ soma size at E18.5 in control and Prox1cKO/cKO mice. t(4) = 6.072, P = 0.0037, unpaired t test, n = 3. (m), Proportion of NPY+ cells at E14.5 and E18.5 in control and Prox1cKO/cKO mice. E14.5: t(4) = 0.2445, P = 0.8189, unpaired t test, n=3; E18.5: t(4) = 0.1160, P = 0.9132, unpaired t test, n=3, All data are presented as mean ± s.e.m.). (n‑p) Artificial exposure to growth factors for 24 h induces precocious RET expression in explanted ganglia of Wnt1‑Cre;R26RTOMATO mice. Triple immunohistochemistry for RET (n) or CASPASE-3 (o) and TH and DAPI on SG sections from P1 Wnt1-Cre;R26RTOMATO mice. The number of RET+ but not CASPASE-3+ cells increases after 24h treatment with ARTN or NRTN (F(2, 8) = 2,782, P = 0.1210, one-way analysis of variance, n=4 for QVD and ARTN and n=3 for NRTN, data are presented as mean ± s.e.m.). (a) and (b) are representative images from experiments carried out on at least two animals. Scale bars: 20 μm in (a-c), (h), (i); 10 μm in (n), (o).
Supplementary Figure 9 A role for Ret in EMN differentiation and for NEM and PEM innervation.
(a, b) NEM and PEM innervation is compromised in Ret-deficient mice. Triple immunohistochemistry for ASMACy3, TH and K5 on NEM sections (a) and for ASMACy3 and TH in PEM sections (b) from P11 control and mutant mice. NEM and PEM of Ret-deficient mice display decreased innervation, compared to control animals. (c) Deletion of Ret in the noradrenergic, but not cholinergic, sympathetic lineage was achieved by driving Cre from the TrkA locus. Triple immunohistochemistry for RET, GFP (recapitulating Ret expression) and TRKA on SG sections from P11 control Retfl/CFP and mutant TrkACre/+;Retfl/CFP mice. RET protein is detectable in RetCFP+TRKA+ EMNs of control (RET+RetCFP+TRKA+ neurons, arrowheads), but not mutant SG neurons (RET-RetCFP+TRKA+ neurons, arrowheads). However, note that RET is still expressed in TRKA- cholinergic neurons (asterisks). (d) Comparison of soma size of NPY+TH+ and CDH8+TH+ (NA4/5) individual neurons (1 dot = 1 neuron) of control Retfl/fl and mutant TrkACre/+;Retfl/fl mice from Fig. 5p confirms that Ret deletion affects soma size of EMN but not of other noradrenergic neurons (NPY+TH+ cells: t(215) = 1.049, P = 0.2954, unpaired t test, n=112 for Retfl/fl and n=105 for TrkACre/+;Retfl/fl; CDH8+TH+ cells: t(267) = 5.588, P < 0.0001, unpaired t test, n=131 for Retfl/fl and n=138 for TrkACre/+;Retfl/fl). (c) are representative images from experiments carried out on at least two animals. Scale bar: 50 μm in (a), 20 μm in (b) and (c).
Supplementary Figure 10 Schematic summary of all results.
The scheme illustrates the initiation and extinction of expression of growth factor receptors and markers of sympathetic neurons, blood vessel mediated path finding of axons to the target region, organogenesis of the NEM and PEM, onset of expression of growth factor ligands in the target erector muscles and concomitant extinction in blood vessels, cell body growth and specialization of NEM and PEM neurons and lastly, innervation of erector muscle target tissues as we interpret our results. Arrows indicate onset (upwards arrow) or extinction (downward arrow) of expression. Note that Ret is expressed in the neuroblasts, which extend axons along blood vessels towards the target, but it is thereafter downregulated in the newly born neurons and it is not turned on again until postnatal stages. Shortly after birth, ARTN is downregulated in blood vessels and upregulated in the nascent erector muscles. Ret re-expression in prospective EMNs participates in erector muscle specialization and target innervation. Abbreviations: BV, blood vessels; NEM, nipple erection muscle; PEM, piloerection muscle.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–13 (PDF 8541 kb)
Supplementary Table 1: Single cell mRNA expression.
The complete transcriptome is reported for each single cell. Values are transcript molecules counts estimated by using UMI. The assigned cluster (i.e. NA1, NA2, etc) and cell areas of single cells are also indicated. (XLS 26293 kb)
Supplementary Table 2: Complete expression patterns.
For each detected gene the average number of detected mRNA molecules expressed by each of the neuronal types is indicated. (XLSX 686 kb)
Supplementary Table 3: Markers for sympathetic neurons.
List of genes over-expressed in each population. Maximum a posteriori probability (MAP) estimates of the average number of molecules per cell are indicated for each population and baseline. (XLSX 32 kb)
Rights and permissions
About this article
Cite this article
Furlan, A., La Manno, G., Lübke, M. et al. Visceral motor neuron diversity delineates a cellular basis for nipple- and pilo-erection muscle control. Nat Neurosci 19, 1331–1340 (2016). https://doi.org/10.1038/nn.4376
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4376
This article is cited by
-
The sympathetic nervous system arose in the earliest vertebrates
Nature (2024)
-
Local and systemic mechanisms that control the hair follicle stem cell niche
Nature Reviews Molecular Cell Biology (2024)
-
CHIT1-positive microglia act as culprits for spinal motor neuron aging
Science China Life Sciences (2024)
-
CHIT1-positive microglia drive motor neuron ageing in the primate spinal cord
Nature (2023)
-
Sympathetic axonal sprouting induces changes in macrophage populations and protects against pancreatic cancer
Nature Communications (2022)