Abstract
Myelination is essential for nervous system function. Schwann cells interact with neurons and the basal lamina to myelinate axons using known receptors, signals and transcription factors. In contrast, the transcriptional control of axonal sorting and the role of mechanotransduction in myelination are largely unknown. Yap and Taz are effectors of the Hippo pathway that integrate chemical and mechanical signals in cells. We describe a previously unknown role for the Hippo pathway in myelination. Using conditional mutagenesis in mice, we show that Taz is required in Schwann cells for radial sorting and myelination and that Yap is redundant with Taz. Yap and Taz are activated in Schwann cells by mechanical stimuli and regulate Schwann cell proliferation and transcription of basal lamina receptor genes, both necessary for radial sorting of axons and subsequent myelination. These data link transcriptional effectors of the Hippo pathway and of mechanotransduction to myelin formation in Schwann cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Zhao, B. et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 (2008).
Zhang, H. et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J. Biol. Chem. 284, 13355–13362 (2009).
Webster, H.D., Martin, R. & O'Connell, M.F. The relationships between interphase Schwann cells and axons before myelination: a quantitative electron microscopic study. Dev. Biol. 32, 401–416 (1973).
Jessen, K.R., Mirsky, R. & Lloyd, A.C. Schwann cells: development and role in nerve repair. Cold Spring Harb. Perspect. Biol. 7, a020487 (2015).
Svaren, J. & Meijer, D. The molecular machinery of myelin gene transcription in Schwann cells. Glia 56, 1541–1551 (2008).
Feltri, M.L., Poitelon, Y. & Previtali, S.C. How Schwann cells sort axons: new concepts. Neuroscientist 22, 252–265 (2015).
Halder, G., Dupont, S. & Piccolo, S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 13, 591–600 (2012).
Taveggia, C. et al. Neuregulin-1 type III determines the ensheathment fate of axons. Neuron 47, 681–694 (2005).
Engler, A.J., Sen, S., Sweeney, H.L. & Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006).
Reginensi, A. et al. Yap- and Cdc42-dependent nephrogenesis and morphogenesis during mouse kidney development. PLoS Genet. 9, e1003380 (2013).
Feltri, M.L. et al. P0-Cre transgenic mice for inactivation of adhesion molecules in Schwann cells. Ann. NY Acad. Sci. 883, 116–123 (1999).
Feltri, M.L. et al. Conditional disruption of β1 integrin in Schwann cells impedes interactions with axons. J. Cell Biol. 156, 199–209 (2002).
Benninger, Y. et al. Essential and distinct roles for Cdc42 and Rac1 in the regulation of Schwann cell biology during peripheral nervous system development. J. Cell Biol. 177, 1051–1061 (2007).
Grinspan, J.B., Marchionni, M.A., Reeves, M., Coulaloglou, M. & Scherer, S.S. Axonal interactions regulate Schwann cell apoptosis in developing peripheral nerve: neuregulin receptors and the role of neuregulins. J. Neurosci. 16, 6107–6118 (1996).
Vassilev, A., Kaneko, K.J., Shu, H., Zhao, Y. & DePamphilis, M.L. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 15, 1229–1241 (2001).
Hung, H.A., Sun, G., Keles, S. & Svaren, J. Dynamic regulation of Schwann cell enhancers after peripheral nerve injury. J. Biol. Chem. 290, 6937–6950 (2015).
Lopez-Anido, C. et al. Differential Sox10 genomic occupancy in myelinating glia. Glia 63, 1897–1914 (2015).
Garratt, A.N., Voiculescu, O., Topilko, P., Charnay, P. & Birchmeier, C. A dual role of erbB2 in myelination and in expansion of the schwann cell precursor pool. J. Cell Biol. 148, 1035–1046 (2000).
Guo, L., Moon, C., Zheng, Y. & Ratner, N. Cdc42 regulates Schwann cell radial sorting and myelin sheath folding through NF2/merlin-dependent and independent signaling. Glia 61, 1906–1921 (2013).
Topilko, P. et al. Krox-20 controls myelination in the peripheral nervous system. Nature 371, 796–799 (1994).
Kuhlbrodt, K., Herbarth, B., Sock, E., Hermans-Borgmeyer, I. & Wegner, M. Sox10, a novel transcriptional modulator in glial cells. J. Neurosci. 18, 237–250 (1998).
Liu-Chittenden, Y. et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 26, 1300–1305 (2012).
Pellegatta, M. et al. α6β1 and α7β1 integrins are required in Schwann cells to sort axons. J. Neurosci. 33, 17995–18007 (2013).
Srinivasan, R. et al. Genome-wide analysis of EGR2/SOX10 binding in myelinating peripheral nerve. Nucleic Acids Res. 40, 6449–6460 (2012).
Anbanandam, A. et al. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc. Natl. Acad. Sci. USA 103, 17225–17230 (2006).
van der Flier, A. & Sonnenberg, A. Function and interactions of integrins. Cell Tissue Res. 305, 285–298 (2001).
Previtali, S.C. et al. Expression of laminin receptors in Schwann cell differentiation: evidence for distinct roles. J. Neurosci. 23, 5520–5530 (2003).
Berti, C. et al. Nonredundant function of dystroglycan and β1 integrins in radial sorting of axons. Development 138, 4025–4037 (2011).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Horton, J.D. et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA 100, 12027–12032 (2003).
Leblanc, S.E. et al. Regulation of cholesterol/lipid biosynthetic genes by Egr2/Krox20 during peripheral nerve myelination. J. Neurochem. 93, 737–748 (2005).
Le, N. et al. Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of Schwann cell differentiation and myelination. Proc. Natl. Acad. Sci. USA 102, 2596–2601 (2005).
Chang, C. et al. A laminin 511 matrix is regulated by TAZ and functions as the ligand for the α6Bβ1 integrin to sustain breast cancer stem cells. Genes Dev. 29, 1–6 (2015).
Lopez-Fagundo, C., Bar-Kochba, E., Livi, L.L., Hoffman-Kim, D. & Franck, C. Three-dimensional traction forces of Schwann cells on compliant substrates. J. R. Soc. Interface 11, 20140247 (2014).
Jagielska, A. et al. Mechanical environment modulates biological properties of oligodendrocyte progenitor cells. Stem Cells Dev. 21, 2905–2914 (2012).
Irvine, K.D. & Harvey, K.F. Control of organ growth by patterning and Hippo signaling in Drosophila. Cold Spring Harb. Perspect. Biol. 7, a019224 (2015).
Zanconato, F. et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 17, 1218–1227 (2015).
Guo, L. & Teng, L. YAP/TAZ for cancer therapy: opportunities and challenges (review). Int. J. Oncol. 46, 1444–1452 (2015).
Kim, M., Kim, T., Johnson, R.L. & Lim, D.S. Transcriptional co-repressor function of the Hippo pathway transducers YAP and TAZ. Cell Rep. 11, 270–282 (2015).
Aragona, M. et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154, 1047–1059 (2013).
Anliker, B. et al. Lysophosphatidic acid (LPA) and its receptor, LPA1, influence embryonic schwann cell migration, myelination, and cell-to-axon segregation. Glia 61, 2009–2022 (2013).
Grigoryan, T. et al. Wnt/Rspondin/β-catenin signals control axonal sorting and lineage progression in Schwann cell development. Proc. Natl. Acad. Sci. USA 110, 18174–18179 (2013).
Petersen, S.C. et al. The adhesion GPCR GPR126 has distinct, domain-dependent functions in Schwann cell development mediated by interaction with laminin-211. Neuron 85, 755–769 (2015).
Chen, Z.L. & Strickland, S. Laminin γ1 is critical for Schwann cell differentiation, axon myelination, and regeneration in the peripheral nerve. J. Cell Biol. 163, 889–899 (2003).
McKee, K.K. et al. Schwann cell myelination requires integration of laminin activities. J. Cell Sci. 125, 4609–4619 (2012).
Grove, M. & Brophy, P.J. FAK is required for Schwann cell spreading on immature basal lamina to coordinate the radial sorting of peripheral axons with myelination. J. Neurosci. 34, 13422–13434 (2014).
Schwartz, M.A. Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb. Perspect. Biol. 2, a005066 (2010).
Paavola, K.J., Sidik, H., Zuchero, J.B., Eckart, M. & Talbot, W.S. Type IV collagen is an activating ligand for the adhesion G protein-coupled receptor GPR126. Sci. Signal. 7, ra76 (2014).
Chance, P.F. et al. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell 72, 143–151 (1993).
Quattrini, A. et al. Beta 4 integrin and other Schwann cell markers in axonal neuropathy. Glia 17, 294–306 (1996).
Gokey, N.G., Srinivasan, R., Lopez-Anido, C., Krueger, C. & Svaren, J. Developmental regulation of microRNA expression in Schwann cells. Mol. Cell. Biol. 32, 558–568 (2012).
Colombelli, C. et al. Perlecan is recruited by dystroglycan to nodes of Ranvier and binds the clustering molecule gliomedin. J. Cell Biol. 208, 313–329 (2015).
Tse, J.R. & Engler, A.J. Preparation of hydrogel substrates with tunable mechanical properties. Curr. Protoc. Cell Biol. 10, 10.16 (2010).10.1002/0471143030.cb1016s47
Zhao, R., Chen, C.S. & Reich, D.H. Force-driven evolution of mesoscale structure in engineered 3D microtissues and the modulation of tissue stiffening. Biomaterials 35, 5056–5064 (2014).
Toda, K., Small, J.A., Goda, S. & Quarles, R.H. Biochemical and cellular properties of three immortalized Schwann cell lines expressing different levels of the myelin-associated glycoprotein. J. Neurochem. 63, 1646–1657 (1994).
Jones, E.A. et al. Distal enhancers upstream of the Charcot-Marie-Tooth type 1A disease gene PMP22. Hum. Mol. Genet. 21, 1581–1591 (2012).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Robinson, M.D., McCarthy, D.J. & Smyth, G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Acknowledgements
We thank E. Hurley for technical assistance; A. Sonnenberg (Netherlands Cancer Institute), D. Meijer and P. Brophy (Centre for Neuroregeneration, Edinburgh), the late G. Tarone (University of Turin), L. Sorokin (University of Muenster) and M. Wegner (Friedrich-Alexander University Erlangen–Nürnberg) for antibodies, and the late R. Quarles (National Institute of Neurological Diseases and Stroke) for the S16 cells. This work was funded by grants NS045630 (to M.L.F.), NS096104 (to L.W.) and NS075269 (to J.S.).
Author information
Authors and Affiliations
Contributions
Y.P., K.C., C.B., M.P. and M.L.F. designed research and interpreted data; Y.P. performed experiments with assistance from C.L.-A., K.C., C.B., M.P., C.W., D.A., K.A. and Y.H.; C.L.-A. and J.S. designed and performed ChIP sequencing and promoter analysis. M.A. and R.Z. designed and helped to perform biomechanical experiments; A.G. and J.L.W. and L.W. contributed analytical tools; F.J.S. analyzed RNA-seq data; Y.P. and M.L.F. wrote the manuscript; Y.P., C.L.-A., R.Z., F.J.S., J.S., L.W. and M.L.F. analyzed data and critically reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
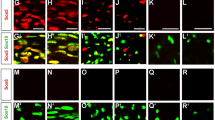
Supplementary Figure 1 Schwann cell proliferation and apoptosis are not affected by Taz or Yap ablation at P20.
(a) TUNEL staining (red), pH3 staining (green) and DAPI (blue) analysis on longitudinal section of sciatic nerves from control, Taz and Yap mutants at P20. Scale bar, 100 µm. At least three animals per genotypes were analyzed. (b) Relative number of TUNEL and pH3 positive nuclei, density of nuclei (number of nuclei per mm2 of sciatic nerve) and total number of nuclei in sciatic nerve (length of sciatic nerve measured: 400 μm). The number of Schwann cells is decreased in double mutants. The increase of nuclei density in Taz cKO; Yap cHet and Yap/Taz cKO is likely caused by absence of myelin. n = 6 control mice, 3 Taz cKO, 4 Yap cKO, 4 Taz cKO Yap cHet, 5 Yap cKO Taz cHet; 3 Taz and Yap cKO. One way ANOVA TUNEL P = 0.4768 F (5, 15) = 0.9519 Taz cKO Yap cHet P = 0.1125; One way ANOVA nuclei per mm2 P < 0.0001, F (5, 18) = 18.54 with Bonferroni post hoc test Taz cKO P = 0.2804; Taz cKO Yap cHet P = 0.0002, Taz and Yap cKO Yap cHet P = 0.0062. Two-tailed unpaired Student's t test Taz and Yap cKO nuclei number P = 0.05. Error bars indicate s.e.m. * P < 0.05, ** P < 0.01, *** P < 0.001.
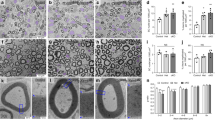
Supplementary Figure 2 The phenotypes of Yap and Taz cKO mice are not due to reduced expression of ErbB2, Cdc42, Egr2 or Sox10 in vivo.
(a) H3K27ac ChIP-Seq enrichment profiles in P15 rat sciatic nerve near (i) Erbb2, (ii) Cdc42, (iii) Egr2 and (iv) Sox10. H3K27ac regions were used to identify the presence of Tead motifs (vertical black bars). (b) Relative mRNA levels in primary rat Schwann cells treated with Verteporfin. Expression of ErbB2, Cdc42 and Sox10 are decreased upon Verteprofin treatment. Error bars indicate s.d. n = 3 independent experiments. Two way ANOVA P < 0.0001, F (2, 24) = 40.96 with Bonferroni post hoc test Erbb2 2 µM P = 0.0136, Erbb2 10 µM P < 0.0001, Cdc42 2 µM P = 0.2804, Cdc42 10 µM P < 0.0001, Egr2 2 µM P = 0.0134, Egr2 10 µM P < 0.0001, Sox10 2 µM P = 0.2312, Sox10 10 µM P < 0.0001. * P < 0.05, **** P < 0.0001. A logarithmic scale was used the y-axis and the origin was set to 1. (c-d) mRNA c) and protein d) levels in control, Taz cKO and Taz cKO; Yap cHet. Expression of Cdc42, ErbB2, Egr2 or Sox10 are not affected in vivo in the mutants. Error bars indicate s.e.m. n=3 (animal). n = 3 mice; One way ANOVA Cdc42 P = 0.3437, Dag1 F (2, 6) = 1.283 with Bonferroni post hoc test Taz cKO Yap cHet P = 0.3454; One way ANOVA ErbB2 P = 0.4162, F (2, 6) = 1.012 with Bonferroni post hoc test Taz cKO Yap cHet P = 0.476. A logarithmic scale was used the y-axis and the origin was set to 1. The western blots were cropped and the complete blots are presented in Supplementary Figure 4.
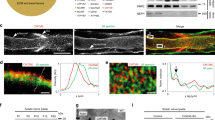
Supplementary Figure 3 Itga6 is regulated by Yap and Taz with Tead.
(a) H3K27ac ChIP-Seq enrichment profiles in P15 rat sciatic nerve near (i) Itga6 and (ii) Dag1. H3K27ac regions were used to identify the presence of Tead motifs (vertical black bars). (b) ChIP-qPCR analysis on S16 Schwann cells. Enrichments are compared to goat IgG. The negative control site for ChIP-qPCR is a region 17.8 kb from the Tekt3 gene, which is not expressed in Schwann cells. Error bars indicate s.d. n=3 n = 3 independent experiments. Two way ANOVA P < 0.0001 F (2, 42) = 37.94 with Bonferroni post hoc test Sox10 – 20 kb P < 0.0001, Tead1 – 20 kb P < 0.0001, Sox10 – 7.6 kb P = 0.0026, Sox10 + 30 bp P < 0.0001. ** P < 0.01, **** P < 0.0001.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 (PDF 1271 kb)
Supplementary Table 1
Genes differentially expressed in Taz cKO–Yap cHet with a false discovery rate of 5% or lower. (XLSX 206 kb)
Rights and permissions
About this article
Cite this article
Poitelon, Y., Lopez-Anido, C., Catignas, K. et al. YAP and TAZ control peripheral myelination and the expression of laminin receptors in Schwann cells. Nat Neurosci 19, 879–887 (2016). https://doi.org/10.1038/nn.4316
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4316
This article is cited by
-
Cellular mechanisms of heterogeneity in NF2-mutant schwannoma
Nature Communications (2023)
-
Hydrogen peroxide induced by nerve injury promotes axon regeneration via connective tissue growth factor
Acta Neuropathologica Communications (2022)
-
Astrocytic YAP prevents the demyelination through promoting expression of cholesterol synthesis genes in experimental autoimmune encephalomyelitis
Cell Death & Disease (2021)
-
Gene expression profiles of YAP1, TAZ, CRB3, and VDR in familial and sporadic multiple sclerosis among an Iranian population
Scientific Reports (2021)
-
Myelin Biology
Neurotherapeutics (2021)