Abstract
EGFRvIII-STAT3 signaling is important in glioblastoma pathogenesis. Here, we identified the cytokine receptor OSMR as a direct target gene of the transcription factor STAT3 in mouse astrocytes and human brain tumor stem cells (BTSCs). We found that OSMR functioned as an essential co-receptor for EGFRvIII. OSMR formed a physical complex with EGFRvIII, and depletion of OSMR impaired EGFRvIII-STAT3 signaling. Conversely, pharmacological inhibition of EGFRvIII phosphorylation inhibited the EGFRvIII-OSMR interaction and activation of STAT3. EGFRvIII-OSMR signaling in tumors operated constitutively, whereas EGFR-OSMR signaling in nontumor cells was synergistically activated by the ligands EGF and OSM. Finally, knockdown of OSMR strongly suppressed cell proliferation and tumor growth of mouse glioblastoma cells and human BTSC xenografts in mice, and prolonged the lifespan of these mice. Our findings identify OSMR as a critical regulator of glioblastoma tumor growth that orchestrates a feed-forward signaling mechanism with EGFRvIII and STAT3 to drive tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005).
Robins, H.I., Chang, S., Butowski, N. & Mehta, M. Therapeutic advances for glioblastoma multiforme: current status and future prospects. Curr. Oncol. Rep. 9, 66–70 (2007).
Maher, E.A. et al. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 15, 1311–1333 (2001).
Furnari, F.B. et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 21, 2683–2710 (2007).
Wen, P.Y. & Kesari, S. Malignant gliomas in adults. N. Engl. J. Med. 359, 492–507 (2008).
Holland, E.C. Gliomagenesis: genetic alterations and mouse models. Nat. Rev. Genet. 2, 120–129 (2001).
Bachoo, R.M. et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 1, 269–277 (2002).
Uhrbom, L. et al. Ink4a-Arf loss cooperates with KRas activation in astrocytes and neural progenitors to generate glioblastomas of various morphologies depending on activated Akt. Cancer Res. 62, 5551–5558 (2002).
Bajenaru, M.L. et al. Optic nerve glioma in mice requires astrocyte Nf1 gene inactivation and Nf1 brain heterozygosity. Cancer Res. 63, 8573–8577 (2003).
Louis, D.N. Molecular pathology of malignant gliomas. Annu. Rev. Pathol. 1, 97–117 (2006).
Singh, S.K. et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 63, 5821–5828 (2003).
Singh, S.K. et al. Identification of human brain tumour initiating cells. Nature 432, 396–401 (2004).
Galli, R. et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 64, 7011–7021 (2004).
Yuan, X. et al. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene 23, 9392–9400 (2004).
Wong, A.J. et al. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc. Natl. Acad. Sci. USA 89, 2965–2969 (1992).
Nagane, M., Lin, H., Cavenee, W.K. & Huang, H.J. Aberrant receptor signaling in human malignant gliomas: mechanisms and therapeutic implications. Cancer Lett. 162, S17–S21 (2001).
de la Iglesia, N. et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 22, 449–462 (2008).
de la Iglesia, N., Puram, S.V. & Bonni, A. STAT3 regulation of glioblastoma pathogenesis. Curr. Mol. Med. 9, 580–590 (2009).
Mosley, B. et al. Dual oncostatin M (OSM) receptors. Cloning and characterization of an alternative signaling subunit conferring OSM-specific receptor activation. J. Biol. Chem. 271, 32635–32643 (1996).
Inda, M.M. et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 24, 1731–1745 (2010).
Fan, Q.W. et al. EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell 24, 438–449 (2013).
Biernat, W., Huang, H., Yokoo, H., Kleihues, P. & Ohgaki, H. Predominant expression of mutant EGFR (EGFRvIII) is rare in primary glioblastomas. Brain Pathol. 14, 131–136 (2004).
Zadeh, G., Bhat, K.P. & Aldape, K. EGFR and EGFRvIII in glioblastoma: partners in crime. Cancer Cell 24, 403–404 (2013).
de la Iglesia, N. et al. Deregulation of a STAT3-interleukin 8 signaling pathway promotes human glioblastoma cell proliferation and invasiveness. J. Neurosci. 28, 5870–5878 (2008).
Bonni, A. et al. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science 278, 477–483 (1997).
Jahani-Asl, A. & Bonni, A. iNOS: a potential therapeutic target for malignant glioma. Curr. Mol. Med. 13, 1241–1249 (2013).
Puram, S.V. et al. STAT3-iNOS Signaling Mediates EGFRvIII-Induced Glial Proliferation and Transformation. J. Neurosci. 32, 7806–7818 (2012).
Caffarel, M.M. & Coleman, N. Oncostatin M receptor is a novel therapeutic target in cervical squamous cell carcinoma. J. Pathol. 232, 386–390 (2014).
Winder, D.M. et al. Overexpression of the oncostatin M receptor in cervical squamous cell carcinoma cells is associated with a pro-angiogenic phenotype and increased cell motility and invasiveness. J. Pathol. 225, 448–462 (2011).
Raz, R., Lee, C.K., Cannizzaro, L.A., d'Eustachio, P. & Levy, D.E. Essential role of STAT3 for embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. USA 96, 2846–2851 (1999).
Kelly, J.J. et al. Proliferation of human glioblastoma stem cells occurs independently of exogenous mitogens. Stem Cells 27, 1722–1733 (2009).
Luchman, H.A. et al. An in vivo patient-derived model of endogenous IDH1-mutant glioma. Neuro. Oncol. 14, 184–191 (2012).
Di Giorgio, F.P., Carrasco, M.A., Siao, M.C., Maniatis, T. & Eggan, K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat. Neurosci. 10, 608–614 (2007).
Nagai, M. et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat. Neurosci. 10, 615–622 (2007).
Bryant, D.M. et al. A molecular network for de novo generation of the apical surface and lumen. Nat. Cell Biol. 12, 1035–1045 (2010).
Huang, W., Sherman, B.T. & Lempicki, R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Soleimani, V.D., Palidwor, G.A., Ramachandran, P., Perkins, T.J. & Rudnicki, M.A. Chromatin tandem affinity purification sequencing. Nat. Protoc. 8, 1525–1534 (2013).
Soleimani, V.D. et al. Transcriptional dominance of Pax7 in adult myogenesis is due to high-affinity recognition of homeodomain motifs. Dev. Cell 22, 1208–1220 (2012).
Soleimani, V.D. et al. Snail regulates MyoD binding-site occupancy to direct enhancer switching and differentiation-specific transcription in myogenesis. Mol. Cell 47, 457–468 (2012).
Bailey, T.L., Williams, N., Misleh, C. & Li, W.W. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373 (2006).
Grant, C.E., Bailey, T.L. & Noble, W.S. FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011).
Bewick, V., Cheek, L. & Ball, J. Statistics review 12: survival analysis. Crit. Care 8, 389–394 (2004).
Lavictoire, S.J., Parolin, D.A., Klimowicz, A.C., Kelly, J.F. & Lorimer, I.A. Interaction of Hsp90 with the nascent form of the mutant epidermal growth factor receptor EGFRvIII. J. Biol. Chem. 278, 5292–5299 (2003).
Acknowledgements
These studies were carried out with support of grants to A.B. from the US National Institutes for Health (NS064007) and the Mathers Foundation, to M.A.R. from the US National Institutes for Health (R01AR044031), the Canadian Institutes for Health Research (CIHR, MOP-81288), and to A.J.-A. from the new investigator startup funds at the LDI/McGill University. M.A.R. is funded as the Canada Research Chair in Molecular Genetics. V.D.S. is funded as the Canada research chair in stem cell epigenetics. A.J.-A., H.Y. and N.C.C. were supported by postdoctoral fellowships from the CIHR. H.A.L. and S.W. are supported by grants from the Alberta Cancer Foundation and the Stem Cell Network. A.M.S. is supported by National Health and Medical Research Council grant and the Operational Infrastructure Support Program provided by the Victorian Government. We thank C. Porter at the Ottawa Hospital Research Institute for critical help with genomic data analyses.
Author information
Authors and Affiliations
Contributions
A.J.-A., M.A.R., and A.B. designed the experiments, analyzed the data and wrote the manuscript. A.J.-A. performed the experiments and analyses. H.Y. and V.D.S. contributed to the analyses of RNA-seq and ChIP-seq data. T.H. contributed to RT-q-PCR, immunoblotting, immunostaining and tissue-culture work. H.A.L. contributed to the characterization of BTSCs and generation of STAT3-kncokdown BTSC73. N.C.C. contributed to PLA analyses and co-immunoprecipitation. M.-C.S. contributed to RT-q-PCR and ChIP-PCR. S.V.P. contributed to STAT3 microarray data in astrocytes. A.M.S. generated mouse EGFRvIII antibody. I.A.J.L. generated rabbit EGFRvIII-antibody. T.J.P. contributed to the bioinformatics analyses of TCGA and Rembrandt data. K.L.L. generated and characterized BTSC 112, 139 and 145 cell lines. S.W. generated and characterized human BTSC12, 25, 30, 41, 68, 73, 90 cell lines.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Gene expression profiling of EGFRvIII-expressing human brain tumor stem cells (BTSCs) and mouse astrocytes
(a) The protein lysates of human BTSCs were analyzed by immunoblotting using the antibodies indicated on the blots. Wild type EGFR and EGFRvIII bands are marked with * and **, respectively. (b) Correlation of EGFRvIII expression with OSMR or phosphorylated STAT3 (p-STAT3-Ser-727) was obtained by running Pearson analyses on the densitometric values of protein expression normalized to tubulin. (c) Three EGFRvIII-expressing BTSCs (# 68, 73, and 90) and a BTSC that did not express EGFRvIII (# 41) were used for gene expression profiling by RNA-seq analyses. Differentially expressed genes in each of the BTSC68, 73, and 90 were called relative to BTSC41 control by Tophat/Cufflinks RNA-seq analysis pipeline. Hierarchical clustering was performed to group genes with similar expression patterns in at least two EGFRvIII-expressing BTSCs relative to control in each pair-wise comparison and the heat map plot was generated to show the expression ratios of the genes that are differentially expressed [fold change (fc) > 5; p value <10−5]. Yellow and blue colors indicate upregulated or downregulated genes in EGFRvIII-expressing lines, respectively. (d) Among candidate target genes that were upregulated in an EGFRvIII dependent manner based on RNA-seq analyses, 10 randomly selected genes and OSMR were validated by RT-q-PCR in EGFRvIII-expressing BTSC73 versus three different BTSC control lines that do not express EGFRvIII (#12, 41, and 139). Gene expression was normalized to GAPDH. The graph represents downregulation of these genes in control lines relative to BTSC73. (e) EGFRvIII-expressing or MSCV-infected control astrocytes that express STAT3 (Stat3loxP/loxP) or in which STAT3 is conditionally deleted (Stat3-/-) were subjected to RNA-seq analyses as described for human BTSC. These are labeled EGFRvIII; Stat3loxP/loxP, EGFRvIII; Stat3-/-, MSCV; Stat3loxP/loxP and MSCV; Stat3-/- on the heat map. The heat map plot (top panel) shows the genes that are downregulated (417 genes, fc ≤ 2) in EGFRvIII; Stat3-/- compared to EGFRvIII; Stat3loxP/loxP. Most of these 417 genes are unchanged between MSCV; Stat3loxP/loxP and MSCV;Stat3-/-. The heat map plot (lower panel) shows the genes that are upregulated (47 genes, fc ≥2) in EGFRvIII; Stat3-/- compared to EGFRvIII; Stat3loxP/loxP. Red color indicates highly expressed genes and white color indicates low expression.
Supplementary Figure 2 Analysis of STAT3 binding sites in EGFRvIII-expressing astrocytes
(a-f) EGFRvIII/Stat3loxP/loxP astrocytes were subjected to ChIP-seq analyses using a STAT3 or an IgG control antibody. 7,725 STAT3 ChIP-seq peaks were called by BeyesPeak with posterior probability equal or more than 0.995. 16,078 negative peaks were called by BeyesPeak and represent genomic regions enriched in IgG control groups compared to STAT3. 7,725 negative peaks with identical length distribution as STAT3 ChIP-seq peaks were randomly selected from the negative peak pool and used as control. Peak height is defined as area under the curve divided by peak length. Averaged conservation scores for STAT3 ChIP-seq peaks were calculated based on phastcon scores (0~1; UCSC 20-way placental mammals) of all the base pairs within each peak groups. Base pairs located within exon features (based on UCSC mm9 refGene table) and repetitive sequences (based on RepBase 14.09) were excluded from the calculation. STAT3 peaks were generated and divided into 100 percentiles in the order of decreasing peak height. (a) The numbers of STAT3 motif variants surrounding centers of STAT3 ChIP-seq peaks is shown. The center of a STAT3 ChIP-seq peak was defined as the base pair located at the middle of the peak. The location of a STAT3 motif was defined as the position of the 5th base pair in the 9nt-long STAT3 motif. The number of each STAT3 motif variants was indicated above each plot within total STAT3 peaks that were binned into 20-bp windows over ±500 bp range of STAT3 peak centers. The expected occurrence of each STAT3 motif variant within the same ±500 bp range of the 7,725 STAT3 peak centers were calculated based on the sequence of each STAT3 motif variant and the background frequencies of each A/C/G/T nucleotide in mouse genome. For each STAT3 motif variant, the p-value from binomial test is shown if the actual occurrence of that STAT3 motif variant is more than the expected occurrence. (b) A logo image of STAT3 motifs in astrocytes identified by MEME in 1,000 STAT3 ChIP-seq peaks is shown. (c) STAT3 peaks and negative peaks were divided into 100 percentiles in the order of decreasing peak height. Average number of STAT3 motifs within each percentile was plotted. “Motif-containing STAT3 peaks” are defined as STAT3 peaks that contain STAT3 motifs. The occurrence of STAT3 motifs was calculated by “Find Individual Motif Occurrences” (FIMO, q-value ≤ 0.1) with the scoring matrix of STAT3 motif identified by Multiple EM for motif Elicitation (MEME) analysis in 1,000 STAT3 ChIP-seq peaks with highest posterior probabilities (p ≤ 1e-4). Notably, negative control peaks are relatively depleted for STAT3 motifs comparing to STAT3 peaks. (d) Conservation score for each peak percentile is plotted. The highest scoring peaks are relatively more conserved. (e) Distribution of STAT3 ChIP-seq peaks over genomic features is shown. Gene annotation is based on UCSC mm9 refGene table. The genomic features are mutually exclusive. In case of overlapping features, numbers of base pairs within STAT3 ChIP-seq peaks were assigned to the genomic feature with the following priority order: promoter/downstream regions > coding exon > 5' UTR > 3' UTR > intron > distal intergenic. Promoter regions were grouped as genomic regions covering -1 kb to transcription start site (TSS), -2kb to -1 kb, -3 kb to -2 kb upstream of TSS and similar three 1-kb windows downstream of TSS. Distal intergenic regions were defined as genomic regions not overlapping with any gene-related features. (f) Distribution of genomic features within mouse genome is shown. The native representation of various genomic features within mouse genome (2,654,895,218 bp) was calculated based on the same method as described for panel e. Comparing to the distribution of STAT3 ChIP-seq peaks, STAT3 predominantly occupies promoter and downstream regions surrounding TSS and is particularly enriched within proximal promoter regions (-1 kb to TSS).
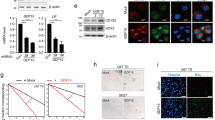
Supplementary Figure 3 OSMR is a direct target gene of STAT3
(a-b) STAT3 Kaplan-Meier survival plots were generated for glioblastoma patients with elevated STAT3 levels using the TCGA or REMBRANDT database, as described in the methods section. (c-d) Multivariate analysis with patient age, IDH mutation status and tumor grade as covariates were performed using two main analytical approaches of Cox proportional hazards (left panel) and Kaplan-Meier stratified logranks (right panel). OSMR and STAT3 are important predictors of survival even after controlling for IDH1 mutation, age, and tumor grade. Abbreviations are: O, OSMR; T, tumor grade; A, age; I, IDH1 mutation; S, STAT3; O|T,A,I: correlation of OSMR upregulation to survival after taking into consideration tumor grade, age, IDH1 mutation; S|T,A,I: correlation of STAT3 upregulation to survival after taking into consideration tumor grade, age, IDH1 mutation as co-variants. (e) STAT3 ChIP-Seq peak obtained at the Osmr promoter is shown. The peak and the STAT3 motif within the peak are both highly conserved across different species. (f) Protein-DNA complexes of EGFRvIII-expressing astrocytes were subjected to immunoprecipitation with a STAT3 antibody or rabbit IgG control. Immunoprecipitated DNA was analyzed by RT-qPCR using two different ChIP primer pairs (OSMR-a, OSMR-b) designed to amplify around the peak summit. An unknown region of chromatin 5 was used as negative control. (g) Lysates of MSCV; Stat3loxP/loxP, EGFRvIII; Stat3loxP/loxP, MSCV; Stat3-/-, and EGFRvIII; Stat3-/- astrocytes were subjected to immunoblotting using OSMR and STAT3 antibodies. Tubulin was used as loading control. (h) mRNA obtained from EGFRvIII; Stat3loxP/loxP and EGFRvIII; Stat3-/- was analyzed by RT-q-PCR to validate 28 randomly selected target genes identified in the RNA-Seq analyses. Fold change was normalized to GAPDH. OSMR is among validated target genes that are highly expressed in EGFRvIII; Stat3loxP/loxP relative to EGFRvIII; Stat3-/- (i) siRNA knockdown of STAT3 was induced in EGFRvIII-expressing human BTSC73 and the RNA of BTSC73-STAT3-KD and corresponding scramble control were subjected to RT-q-PCR analyses using primers for STAT3 and OSMR. Fold change expressions were normalized to GAPDH.
Supplementary Figure 4 EGFRvIII and OSMR form a physical complex independently of gp130
(a-b) EGFRvIII-expressing human BTSCs were subjected to immunostaining using antibodies against EGFRvIII, OSMR, and connexin 43 as indicated on the panels. Nuclei were stained with Hoechst. (c) EGFRvIII; Stat3loxP/loxP astrocytes were subjected to immunoprecipitation using a specific EGFRvIII antibody followed by immunoblotting analyses with a gp130 or pan-EGFR antibody. No interaction of EGFRvIII with gp130 or wild type EGFR (indicated with a * on the blot) was detected. (d-e) MSCV-expressing or EGFRvIII-expressing astrocytes were transfected with siRNAs against gp130 (panel d) or wild type EGFR (panel e) and analyzed by immunoblotting using antibodies indicated on the blot. Tubulin was used as loading control. (f) EGFRvIII-expressing astrocytes transfected with siRNA against gp130, EGFR, or scramble control were subjected to PLA analyses using OSMR and EGFRvIII antibodies as described in Figure 3. (g) EGFRvIII-expressing astrocytes transfected with siRNA against gp130, EGFR, or scramble control were subjected to immunoblotting analyses with phospho-STAT3 antibodies as indicated on the blot. Tubulin was used as loading control.
Supplementary Figure 5 The ligand OSM regulates the phosphorylation of EGFR and STAT3 in wild type astrocytes
(a-b) Stat3loxP/loxP astrocytes were treated with 10 ng/ml of OSM, and the level of activated STAT3 was evaluated by immunohistochemical analyses using the p-STAT3-Y705 (panel a) and p-STAT3-Ser727 (panel b) antibodies. Nuclei were stained with Hoechst. (c) Stat3loxP/loxP and conditional Stat3-/- astrocytes were treated with 10 or 100 ng/ml of OSM, and the level of activated STAT3 was evaluated by immunoblotting. Tubulin was used as loading control. (d) Stat3loxP/loxP mouse astrocytes were treated with 10 ng/ml of OSM or EGF at different time points and lysates were blotted with antibodies indicated on the blot. (e) Stat3loxP/loxP and conditional Stat3-/- astrocytes were treated with 10 and 100 ng/ml of either OSM or EGF or both. Lysates were analyzed by immunoblotting with phospho-STAT3 and EGFR antibodies following 30 minutes of treatment. Tubulin was used as loading control. (f-h) Stat3loxP/loxP astrocytes were treated with 10 ng/ml of OSM and subjected to immunocytochemical analyses using a Ki67 antibody. The percentage of Ki67 positive cells in the absence and presence of OSM is presented in h.
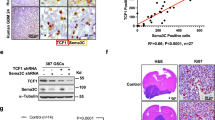
Supplementary Figure 6 Efficient knockdown of OSMR in mouse astrocytes and human BTSCs
(a) EGFRvIII-expressing BTSC73 was targeted by 3 different shRNA spanning the OSMR gene using lentiviral-mediated system as described in the Methods section. Representative panels of untransduced BTSC73 and BTSC73 transduced with either GFP-expressing scramble control or GFP-expressing-OSMR-knockdowns (KD) are shown. The brightest GFP positive cells in BTSC73 were FACS-sorted (right panel) following 1 week in culture. (b) RT-q-PCR analyses of OSMR-KD1, -KD2, and -KD3-BTSC73 and scrambled control-BTSC73 are shown. Gene expression was normalized to GAPDH. (c) Protein lysates of BTSC112 (expressing EGFRvIII and high level of phosphorylated STAT3), BTSC139 (not expressing EGFRvIII or high level of phosphorylated STAT3) and BTSC145 (not expressing EGFRvIII but expressing high level of phosphorylated STAT3) were subjected to immunoblotting analyses using antibodies indicated on the blot. Tubulin was used as loading control. (d) Correlation of phosphorylated STAT3 (Y705) with EGFR (Y1068) was obtained by performing Pearson analyses on clinical glioblastoma samples at TCGA for which proteomic data was available for these phosphorylation sites. (e) EGFRvIII/Stat3loxP/loxP astrocytes were transduced with lentiviruses carrying either short hairpin RNAs (shRNA) against OSMR or a scrambled shRNA control. Transduced cells were selected with blasticidin. mRNA for EGFRvIII-expressing astrocytes that were transduced with either of shOSMR1 and 2 or scrambled shRNA was analyzed by q-RT-PCR using 5 different primer pairs (denoted 1, 2, 3, 4, 5 on the X axis) that were designed against different regions of Osmr gene. Expression was normalized to GAPDH (f) Protein lysates of EGFRvIII-expressing astrocytes were subjected to immunoblotting analyses using the OSMR antibody in the absence and presence of an OSMR blocking peptide (top panel) to confirm the specificity of the OSMR antibody. Tubulin was used as loading control. Lysates of OSMR-KD1, -KD2 and scrambled control EGFRvIII-expressing astrocytes were subjected to immunoblotting analyses using an OSMR antibody. (bottom panel). Tubulin was used as loading control. (g) OSMR-KD1, -KD2 and scrambled control were subjected to immunostaining with an antibody against OSMR and Hoechst. (h) OSMR-KD1, -KD2 and scrambled control were subjected to immunostaining with an antibody against ki67 and Hoechst.
Supplementary Figure 7 Full scans of Western blots in the main figures
The full scans of Western blots in the main figures are presented. In some cases the membranes were cut into different sections for blotting with proteins of different MW, to avoid re-probing or stripping the membranes or running multiple gels for the same samples.
Supplementary Figure 8 Full scans of Western blots for the supplementary figures
The full scans of Western blots for supplementary figures are presented. In some cases, the membranes were cut into different sections for blotting with proteins of different MW, to avoid re-probing or stripping the membranes.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Tables 1–3 (PDF 2635 kb)
Supplementary Methods Checklist
(PDF 452 kb)
Supplementary Table 4
Functional annotation of EGFRvIII common candidate target genes by DAVID Bioinformatics Resources 6.7. (XLS 176 kb)
Rights and permissions
About this article
Cite this article
Jahani-Asl, A., Yin, H., Soleimani, V. et al. Control of glioblastoma tumorigenesis by feed-forward cytokine signaling. Nat Neurosci 19, 798–806 (2016). https://doi.org/10.1038/nn.4295
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4295
This article is cited by
-
Cloning, expression, purification, and immunoblotting analysis of recombinant type III fibronectin domains of human oncostatin M receptor
Molecular Biology Reports (2023)
-
EGFR ligand shifts the role of EGFR from oncogene to tumour suppressor in EGFR-amplified glioblastoma by suppressing invasion through BIN3 upregulation
Nature Cell Biology (2022)
-
Annexin A2–STAT3–Oncostatin M receptor axis drives phenotypic and mesenchymal changes in glioblastoma
Acta Neuropathologica Communications (2020)
-
A STAT3-based gene signature stratifies glioma patients for targeted therapy
Nature Communications (2019)
-
FHL2 interacts with EGFR to promote glioblastoma growth
Oncogene (2018)