Abstract
C9orf72 mutations are the most common cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Dipeptide repeat proteins (DPRs) produced by unconventional translation of the C9orf72 repeat expansions cause neurodegeneration in cell culture and in animal models. We performed two unbiased screens in Saccharomyces cerevisiae and identified potent modifiers of DPR toxicity, including karyopherins and effectors of Ran-mediated nucleocytoplasmic transport, providing insight into potential disease mechanisms and therapeutic targets.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
DeJesus-Hernandez, M. et al. Neuron 72, 245–256 (2011).
Renton, A.E. et al. Neuron 72, 257–268 (2011).
Ash, P.E. et al. Neuron 77, 639–646 (2013).
Mori, K. et al. Science 339, 1335–1338 (2013).
Zu, T. et al. Proc. Natl. Acad. Sci. USA 110, E4968–E4977 (2013).
Mizielinska, S. et al. Science 345, 1192–1194 (2014).
Kwon, I. et al. Science 345, 1139–1145 (2014).
Zhang, Y.J. et al. Acta Neuropathol. 128, 505–524 (2014).
May, S. et al. Acta Neuropathol. 128, 485–503 (2014).
Yamakawa, M. et al. Hum. Mol. Genet. 24, 1630–1645 (2015).
Wen, X. et al. Neuron 84, 1213–1225 (2014).
Elden, A.C. et al. Nature 466, 1069–1075 (2010).
Sun, Z. et al. PLoS Biol. 9, e1000614 (2011).
Armakola, M. et al. Nat. Genet. 44, 1302–1309 (2012).
Krebber, H., Taura, T., Lee, M.S. & Silver, P.A. Genes Dev. 13, 1994–2004 (1999).
Dormann, D. et al. EMBO J. 29, 2841–2857 (2010).
Neumann, M. et al. Brain 134, 2595–2609 (2011).
Bardy, C. et al. Proc. Natl. Acad. Sci. USA 112, E2725–E2734 (2015).
Chew, J. et al. Science 348, 1151–1154 (2015).
Haeusler, A.R. et al. Nature 507, 195–200 (2014).
Hu, Y. et al. Genome Res. 17, 536–543 (2007).
Tong, A.H. et al. Science 294, 2364–2368 (2001).
Collins, S.R., Schuldiner, M., Krogan, N.J. & Weissman, J.S. Genome Biol. 7, R63 (2006).
Huang, D.W., Sherman, B.T. & Lempicki, R.A. Nat. Protoc. 4, 44–57 (2009).
Lagier-Tourenne, C. et al. Proc. Natl. Acad. Sci. USA 110, E4530–E4539 (2013).
Ladewig, J. et al. Nat. Methods 9, 575–578 (2012).
Mertens, J. et al. Am. J. Pathol. 182, 1769–1779 (2013).
Acknowledgements
We thank J. Ravits and C. Lagier-Tourenne (University of California, San Diego) for sharing the control and C9orf72 ALS patient fibroblasts used in this study. We acknowledge microscopy assistance from the Stanford Neuroscience Microscopy Service, supported by US National Institutes of Health (NIH) grant NS069375. This work was supported by NIH grants 1R01NS065317 and 1R01NS073660 (A.D.G.). A.D.G. is supported by the Packard Center for ALS Research at Johns Hopkins and Target ALS. A.J. is supported by the Dean's Postdoctoral Fellowship from Stanford University. S.S. is a recipient of NIH K99/R00 Award (NS091538) and Target ALS Springboard Fellowship. J.M. and F.H.G. are supported by JPB Foundation, the Helmsley Foundation and the Mathers Foundation. Additional research funding was provided by the KU Leuven, the European Research Council in the context of the European's Seventh Framework Programme (FP7/2007-2013 and ERC grant agreement 340429), the Fund for Scientific Research Flanders (FWO-Vlaanderen) G.0983.14N, the Interuniversity Attraction Poles Programme P7/16 initiated by the Belgian Science Policy Office, the Association Belge contre les Maladies Neuro-Musculaires (ABMM), the ALS Liga (Belgium) and the Opening the Future fund. S.B. received a fellowship from the Agency for Innovation by Science and Technology IWT. E.B. holds a postdoctoral fellowship from FWO-Vlaanderen. W.R. is supported through the E. von Behring Chair for Neuromuscular and Neurodegenerative Disorders.
Author information
Authors and Affiliations
Contributions
A.J. designed and performed the yeast experiments with help from N.C., S.B.Y., J.W.P., G.B. and N.J.K. S.B. and E.B. generated and characterized some yeast expression plasmids with direction from L.V.D.B. and W.R. A.J. performed the mouse primary neuron experiments. A.J., S.S., J.M. and J.R.H. worked together on the human iN experiments with direction from F.H.G. A.J. and A.D.G. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
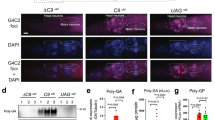
Supplementary Figure 1 C9orf72 DPR localization and effect of modifier genes on localization and protein levels.
a) Cellular localization and expression of C9orf72 DPRs was evaluated by immunocytochemistry. DPRs were visualized using anti-FLAG antibody. b) Upregulation of four karyopherin genes, KAP104, KAP114, KAP120 or KAP122, in yeast does not alter localization of (Pro-Arg)50 DPR. c) (Pro-Arg)50 expression levels were evaluated in deletion strains that mitigate (Pro-Arg)50 toxicity and under conditions of KAP104, KAP114, KAP120 and KAP122 overexpression. dhh1Δ, gtr1Δ, sgo1Δ, ski8Δ, stp1Δ and uaf30Δ lowered levels of (Pro-Arg)50. d) Deletion of these 6 genes did not strongly lower levels of YFP expressed under the same promoter, indicating that their effect on (Pro-Arg)50 levels was likely specific.
Supplementary Figure 2 Effect of (Pro-Arg)50 toxicity modifiers on TDP-43 and α-synuclein (α-syn) toxicity in yeast.
a) Gene deletions that suppressed (Pro-Arg)50 toxicity were tested for unspecific effect on survival by introducing TDP-43 into these deletions. Only ubr2Δ and ski8Δ showed protective effect against TDP43 toxicity in yeast, while others did not. b) ski8Δ did not suppress α-syn toxicity. ubr2Δ suppressed toxicity of all three proteins and therefore this deletion does not confer any specific protection against (Pro-Arg)50 or TDP-43 toxicity, but might represent a common target for all three aggregation-prone proteins.
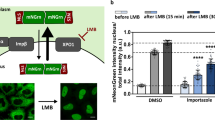
Supplementary Figure 3 KPNA3 upregulation does not affect localization of (Pro-Arg)50 in mouse primary cells.
Cells were transfected with (Pro-Arg)50 and GFP or KPNA3 (C-terminally V5 tagged). Neurons were stained with anti-FLAG antibody and anti-V5, were applicable. Representative images are shown.
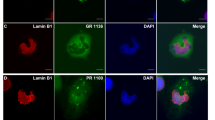
Supplementary Figure 4 Effect of C9orf72 mutation on localization of proteins implicated in nucleocytoplasmic transport.
iNeurons from control subjects or C9orf72 mutation carriers were used to examine localization of a) LmnB and TNPO3, b) RanGAP1 and KPNA3, c) XPO5. We did not observe significant differences in the localization of these proteins in the control cells compared to C9orf72 mutation carrier cells.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 777 kb)
Rights and permissions
About this article
Cite this article
Jovičić, A., Mertens, J., Boeynaems, S. et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 18, 1226–1229 (2015). https://doi.org/10.1038/nn.4085
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4085
This article is cited by
-
Nuclear-import receptors as gatekeepers of pathological phase transitions in ALS/FTD
Molecular Neurodegeneration (2024)
-
Repeat length of C9orf72-associated glycine–alanine polypeptides affects their toxicity
Acta Neuropathologica Communications (2023)
-
Looking for answers far away from the soma—the (un)known axonal functions of TDP-43, and their contribution to early NMJ disruption in ALS
Molecular Neurodegeneration (2023)
-
Mutant GGGGCC RNA prevents YY1 from binding to Fuzzy promoter which stimulates Wnt/β-catenin pathway in C9ALS/FTD
Nature Communications (2023)
-
Single-molecule imaging reveals distinct elongation and frameshifting dynamics between frames of expanded RNA repeats in C9ORF72-ALS/FTD
Nature Communications (2023)