Abstract
Pro-opiomelanocortin (POMC)- and agouti-related peptide (AgRP)-expressing neurons of the arcuate nucleus of the hypothalamus (ARC) are oppositely regulated by caloric depletion and coordinately stimulate and inhibit homeostatic satiety, respectively. This bimodality is principally underscored by the antagonistic actions of these ligands at downstream melanocortin-4 receptors (MC4R) in the paraventricular nucleus of the hypothalamus (PVH). Although this population is critical to energy balance, the underlying neural circuitry remains unknown. Using mice expressing Cre recombinase in MC4R neurons, we demonstrate bidirectional control of feeding following real-time activation and inhibition of PVHMC4R neurons and further identify these cells as a functional exponent of ARCAgRP neuron–driven hunger. Moreover, we reveal this function to be mediated by a PVHMC4R→lateral parabrachial nucleus (LPBN) pathway. Activation of this circuit encodes positive valence, but only in calorically depleted mice. Thus, the satiating and appetitive nature of PVHMC4R→LPBN neurons supports the principles of drive reduction and highlights this circuit as a promising target for antiobesity drug development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Berridge, K.C. Motivation concepts in behavioral neuroscience. Physiol. Behav. 81, 179–209 (2004).
Cannon, W.B. The Wisdom of the Body vol. XV (Norton, 1932).
Cowley, M.A. et al. Integration of NPY, AGRP, and melanocortin signals in the hypothalamic paraventricular nucleus: evidence of a cellular basis for the adipostat. Neuron 24, 155–163 (1999).
Sternson, S.M. Hypothalamic survival circuits: blueprints for purposive behaviors. Neuron 77, 810–824 (2013).
Garfield, A.S., Lam, D.D., Marston, O.J., Przydzial, M.J. & Heisler, L.K. Role of central melanocortin pathways in energy homeostasis. Trends Endocrinol. Metab. TEM 20, 203–215 (2009).
Aponte, Y., Atasoy, D. & Sternson, S.M. AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nat. Neurosci. 14, 351–355 (2011).
Atasoy, D., Betley, J.N., Su, H.H. & Sternson, S.M. Deconstruction of a neural circuit for hunger. Nature 488, 172–177 (2012).
Krashes, M.J. et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J. Clin. Invest. 121, 1424–1428 (2011).
Krashes, M.J., Shah, B.P., Koda, S. & Lowell, B.B. Rapid versus delayed stimulation of feeding by the endogenously released AgRP neuron mediators GABA, NPY, and AgRP. Cell Metab. 18, 588–595 (2013).
Zhan, C. et al. Acute and long-term suppression of feeding behavior by POMC neurons in the brainstem and hypothalamus, respectively. J. Neurosci. 33, 3624–3632 (2013).
Balthasar, N. et al. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell 123, 493–505 (2005).
Fan, W., Boston, B.A., Kesterson, R.A., Hruby, V.J. & Cone, R.D. Role of melanocortinergic neurons in feeding and the agouti obesity syndrome. Nature 385, 165–168 (1997).
Huszar, D. et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 88, 131–141 (1997).
Rossi, M. et al. A C-terminal fragment of Agouti-related protein increases feeding and antagonizes the effect of alpha-melanocyte stimulating hormone in vivo. Endocrinology 139, 4428–4431 (1998).
Shah, B.P. et al. MC4R-expressing glutamatergic neurons in the paraventricular hypothalamus regulate feeding and are synaptically connected to the parabrachial nucleus. Proc. Natl. Acad. Sci. USA 111, 13193–13198 (2014).
Small, C.J. et al. Effects of chronic central nervous system administration of agouti-related protein in pair-fed animals. Diabetes 50, 248–254 (2001).
Yaswen, L., Diehl, N., Brennan, M.B. & Hochgeschwender, U. Obesity in the mouse model of pro-opiomelanocortin deficiency responds to peripheral melanocortin. Nat. Med. 5, 1066–1070 (1999).
Vaisse, C., Clement, K., Guy-Grand, B. & Froguel, P. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat. Genet. 20, 113–114 (1998).
Yeo, G.S. et al. A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat. Genet. 20, 111–112 (1998).
Rossi, J. et al. Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metab. 13, 195–204 (2011).
Leibowitz, S.F., Hammer, N.J. & Chang, K. Hypothalamic paraventricular nucleus lesions produce overeating and obesity in the rat. Physiol. Behav. 27, 1031–1040 (1981).
Xi, D., Gandhi, N., Lai, M. & Kublaoui, B.M. Ablation of Sim1 neurons causes obesity through hyperphagia and reduced energy expenditure. PLoS ONE 7, e36453 (2012).
Atasoy, D., Aponte, Y., Su, H.H. & Sternson, S.M.A. FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J. Neurosci. 28, 7025–7030 (2008).
Krashes, M.J. et al. An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature 507, 238–242 (2014).
Betley, J.N., Cao, Z.F., Ritola, K.D. & Sternson, S.M. Parallel, redundant circuit organization for homeostatic control of feeding behavior. Cell 155, 1337–1350 (2013).
Petreanu, L., Huber, D., Sobczyk, A. & Svoboda, K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat. Neurosci. 10, 663–668 (2007).
Wu, Z. et al. An obligate role of oxytocin neurons in diet induced energy expenditure. PLoS ONE 7, e45167 (2012).
Lu, X.Y., Barsh, G.S., Akil, H. & Watson, S.J. Interaction between alpha-melanocyte-stimulating hormone and corticotropin-releasing hormone in the regulation of feeding and hypothalamo-pituitary-adrenal responses. J. Neurosci. 23, 7863–7872 (2003).
Alexander, G.M. et al. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron 63, 27–39 (2009).
Jennings, J.H., Rizzi, G., Stamatakis, A.M., Ung, R.L. & Stuber, G.D. The inhibitory circuit architecture of the lateral hypothalamus orchestrates feeding. Science 341, 1517–1521 (2013).
Jennings, J.H. et al. Visualizing hypothalamic network dynamics for appetitive and consummatory behaviors. Cell 160, 516–527 (2015).
Armbruster, B.N., Li, X., Pausch, M.H., Herlitze, S. & Roth, B.L. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl. Acad. Sci. USA 104, 5163–5168 (2007).
Sutton, A.K. et al. Control of food intake and energy expenditure by nos1 neurons of the paraventricular hypothalamus. J. Neurosci. 34, 15306–15318 (2014).
Garfield, A.S. et al. A parabrachial-hypothalamic cholecystokinin neurocircuit controls counterregulatory responses to hypoglycemia. Cell Metab. 20, 1030–1037 (2014).
Carter, M.E., Soden, M.E., Zweifel, L.S. & Palmiter, R.D. Genetic identification of a neural circuit that suppresses appetite. Nature 503, 111–114 (2013).
Nagai, K. et al. Lesions in the lateral part of the dorsal parabrachial nucleus caused hyperphagia and obesity. J. Clin. Biochem. Nutr. 2, 103–112 (1987).
Wu, Q., Boyle, M.P. & Palmiter, R.D. Loss of GABAergic signaling by AgRP neurons to the parabrachial nucleus leads to starvation. Cell 137, 1225–1234 (2009).
Wu, Q., Clark, M.S. & Palmiter, R.D. Deciphering a neuronal circuit that mediates appetite. Nature 483, 594–597 (2012).
Vong, L. et al. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron 71, 142–154 (2011).
Paues, J., Mackerlova, L. & Blomqvist, A. Expression of melanocortin-4 receptor by rat parabrachial neurons responsive to immune and aversive stimuli. Neuroscience 141, 287–297 (2006).
Stamatakis, A.M. & Stuber, G.D. Activation of lateral habenula inputs to the ventral midbrain promotes behavioral avoidance. Nat. Neurosci. 15, 1105–1107 (2012).
Atasoy, D. et al. A genetically specified connectomics approach applied to long-range feeding regulatory circuits. Nat. Neurosci. 17, 1830–1839 (2014).
Kirchgessner, A.L. & Sclafani, A. PVN-hindbrain pathway involved in the hypothalamic hyperphagia-obesity syndrome. Physiol. Behav. 42, 517–528 (1988).
McCabe, J.T., DeBellis, M. & Leibowitz, S.F. Clonidine-induced feeding: analysis of central sites of action and fiber projections mediating this response. Brain Res. 309, 85–104 (1984).
Stachniak, T.J., Ghosh, A. & Sternson, S.M. Chemogenetic synaptic silencing of neural circuits localizes a hypothalamus→midbrain pathway for feeding behavior. Neuron 82, 797–808 (2014).
Cai, H., Haubensak, W., Anthony, T.E. & Anderson, D.J. Central amygdala PKC-delta(+) neurons mediate the influence of multiple anorexigenic signals. Nat. Neurosci. 17, 1240–1248 (2014).
Carter, M.E., Han, S. & Palmiter, R.D. Parabrachial calcitonin gene-related Peptide neurons mediate conditioned taste aversion. J. Neurosci. 35, 4582–4586 (2015).
Keys, A. Human starvation and its consequences. J. Am. Diet. Assoc. 22, 582–587 (1946).
Hull, C.L. Principles of Behavior: An Introduction to Behavior Theory vol. X, 421–422 (D. Appleton Century, 1943).
Margules, D.L. & Olds, J. Identical “feeding” and “rewarding” systems in the lateral hypothalamus of rats. Science 135, 374–375 (1962).
Tong, Q., Ye, C.P., Jones, J.E., Elmquist, J.K. & Lowell, B.B. Synaptic release of GABA by AgRP neurons is required for normal regulation of energy balance. Nat. Neurosci. 11, 998–1000 (2008).
Song, H. et al. Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc. Natl. Acad. Sci. USA 109, 17531–17536 (2012).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Kim, J.H. et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE 6, e18556 (2011).
Saxena, A. et al. Trehalose-enhanced isolation of neuronal sub-types from adult mouse brain. Biotechniques 52, 381–385 (2012).
Hempel, C.M., Sugino, K. & Nelson, S.B. A manual method for the purification of fluorescently labeled neurons from the mammalian brain. Nat. Protoc. 2, 2924–2929 (2007).
Tang, F. et al. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat. Protoc. 5, 516–535 (2010).
Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Abercrombie, M. Estimation of nuclear population from microtome sections. Anat. Rec. 94, 239–247 (1946).
Liu, T. et al. Fasting activation of AgRP neurons requires NMDA receptors and involves spinogenesis and increased excitatory tone. Neuron 73, 511–522 (2012).
Franklin, K.B.J. & Paxinos, G. The Mouse Brain in Stereotaxic Coordinates 3rd edn. (Academic Press, Elsevier, 2008).
Acknowledgements
The authors gratefully acknowledge the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Mouse Metabolism Core for technical support, V. Petkova and the Beth Israel Deaconess Medical Center Molecular Medicine Core Facility for assistance with quantitative PCR and sample preparation, and D. Morse for the production of the rabies virus. Sequencing and initial data processing were performed at Massachusetts General Hospital's Next-Gen Sequencing Core. Sequencing was supported in part by funding from the Boston Area Diabetes Endocrinology Research Center (BADERC P30 DK057521). This work was supported by the University of Edinburgh Chancellor's Fellowship (A.S.G.); US National Institutes of Health grants to B.B.L. (R01 DK096010, R01 DK089044, R01 DK071051, R01 DK075632, R37 DK053477, BNORC Transgenic Core P30 DK046200, BADERC Transgenic Core P30 DK057521), to M.J.K. (F32 DK089710), to D.P.O. (K08 DK071561) and to J.K.E. (R01 DK088423 and R37 DK0053301); American Heart Association Postdoctoral Fellowship 14POST20100011 to J.N.C.; viral vector production core P30 NS045776 to B.A.T.; and an American Diabetes Association Mentor-Based Fellowship to B.P.S. and B.B.L. This research was supported, in part, by the Intramural Research Program of the NIH, NIDDK (DK075087, DK075088).
Author information
Authors and Affiliations
Contributions
A.S.G., M.J.K., B.P.S. and B.B.L. conceived the studies. A.S.G., M.J.K., C.L. and J.C.M. conducted the studies with assistance from B.P.S., E.W., J.S.S. and D.P.O. Single cell RNA sequencing profiling was conducted by J.N.C. Energy expenditure assays were conducted by O.G. In situ validation of the MC4R-t2a-Cre mouse line was conducted by C.E.L. and J.K.E. Rabies virus was provided by B.A.T. A.S.G., M.J.K. and B.B.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Central Mc4r-t2a-Cre expression
Mc4r-t2a-Cre expression was demarked by a germline R26-loxSTOPlox-tdTomato reporter allele and assayed across the rostral-caudal extent of the murine neuraxis. The neuroanatomical distribution of MC4R::tdTomato expressing neurons was consistent with the endogenous Mc4r expression profile. Abbreviations: BNST, bed nucleus of the stria terminalis; CeM, central amygdaloid nucleus; DMV, dorsomedial nucleus of the vagus; IML, intermediolateral nucleus; LH, lateral hypothalamus; LPBN, lateral parabrachial nucleus; NTS, nucleus of the solitary tract; PVH, paraventricular nucleus of the hypothalamus; VMH, ventromedial nucleus of the hypothalamus. Scale bar = 100 μm.
Supplementary Figure 2 Validation of PVH Mc4r-t2a-Cre expression
Mc4r-t2a-Cre expression within the paraventricular nucleus of the hypothalamus (PVH) was demarked by injection of a cre-dependent AAV8-hSyn-DIO-GFP viral construct. Colocalization of Mc4r-t2a-Cre::GFP expression with endogenous Mc4r mRNA was determined by way of dual immunohistochemistry (for GFP, brown soma) and radioactive in situ hybridization (for Mc4R mRNA, black puncta). Microscopic analysis revealed extensive colocalisation of the two signals. a, scale bar = 500 μm; b, scale bar = 50 μm; c, scale bar = 10 μm.
Supplementary Figure 3 Neuroanatomical location of optic fibers for in vivo optogenetic occlusion studies
Mice used for optogenetic occlusion studies were validated for fiber placement using histological sections. The approximate positions of the fibers is denoted by an X. a-c, Relates to Figure 1g-h. d, Relates to Figure 2d. e, Relates to Figure 2e. Abbreviations: BNST, bed nucleus of the stria terminalis; LH, lateral hypothalamus; LPBN, lateral parabrachial nucleus; PVH, paraventricular nucleus of the hypothalamus.
Supplementary Figure 4 PVHMC4R neurons do not express oxytocin
Fluorescent immunohistological analysis of MC4R-t2a-Cre::tdTomato (red) and endogenous oxytocin (green) expression in the PVH revealed the complete absence of colocalization at all neuroanatomical levels. Abbreviations: 3v, third ventricle. Scale bar = 100 μm.
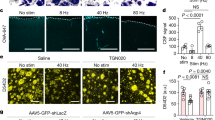
Supplementary Figure 5 Validation of in vivo DREADD expression and function
a-d, Validation of excitatory hM3Dq-mCherry expression in Mc4r-t2a-Cre mice. a, Representative image of hM3Dq-mCherry expression within the PVH. b, Membrane potential and firing rate of Mc4r-t2a-Cre::hM3Dq-mCherryPVH neurons increased upon 5 µM CNO application during electrophysiological current clamp recordings. c, Representative image of hM3Dq-mCherry expression within the BNST. d, Representative image of hM3Dq-mCherry expression within the LH. e-h, Validation of inhibitory hM3Dq-mCherry expression in Mc4r-t2a-Cre mice. e, Representative image of hM4Di-mCherry expression within the PVH. f, Membrane potential and firing rate of Mc4r-t2a-Cre::hM4Di-mCherryPVH neurons decreased upon 5 µM CNO application electrophysiological current clamp recordings. g, Representative image of hM4Di-mCherry expression within the BNST. h, Representative image of hM4Di-mCherry expression within the LH. Abbreviations: 3v, third ventricle; aca, anterior commissure anterior part; f, fornix. Scale bar in a, = 100 μm and relates to all images.
Supplementary Figure 6 PVHMC4R neurons do not influence energy expenditure or locomotor activity, while either PVHOXT or PVHCRH neurons do not influence food intake
a-c, Chemogenetic activation of PVHMC4R neurons did not influence a, locomotor activity (LMA; n=11, Paired two-tailed t-test, t(10)=0.43, p=0.67), b, total energy expenditure (TEE; n=11, Paired two-tailed t-test, t(10)=0.20, p=0.85) or c, respiratory exchange ratio (RER; n=11. Paried two-tailed t-test, t(10)=0.35, p=0.79). d-e, PVHOXT and PVHCRH neurons do not influence feeding behavior. d, Chemogenetic inhibition of PVHOXT neurons did not significantly affect light-cycle food intake, compared to the same mice treated with saline (n= n=6, Repeated measures ANOVA, main effect of treatment and interaction not significant, main effect of time (F(4,25)=70.31, p<0.0001). e, Chemogenetic inhibition of PVHCRH neurons did not significantly affect light-cycle food intake, compared to the same mice treated with saline (n=5, Repeated measures ANOVA, main effect of interaction and treatment not significant, main effect of time (F(3,16)=20.61, p<0.0001)..
Supplementary Figure 7 Efferent targets and connectivity of PVHMC4R projections
Neuroanatomical projection mapping from PVHMC4R neurons was achieved via unilateral stereotaxic injection of a synaptically targeted fluorophore (AAV8-hSyn-FLEX-Syn-mCherry). PVHMC4R neurons exhibit exclusively descending and predominantly ipsilateral projections to the median eminence (ME), retrorubral field (RRF), ventrolateral periaqueductal grey (vlPAG), lateral parabrachial nucleus (LPBN), pre-locus coeruleus (pLC), nucleus of the solitary tract (NTS), dorsal motor nucleus of the vagus (DMV), rostral ventrolateral medulla (RVLM) and intermediolateral nucleus (IML). Qualitative assessment of fiber density demonstrated that the ME, LPBN and NTS received the densest innervation. CRACM analysis: No light-evoked EPSCs were detected on ARCAgRP neurons, confirming the uni-directionality of the ARCAgRP→PVHMC4R circuit. The vlPAG, pLC and NTS exhibited low to no connectivity, 7%, 0% and 5%, respectively. 56% of LPBN and 55% of DMV of randomly patched post-synaptic neurons exhibited light-evoked EPSCs. Abbreviations: 3v, third ventricle; Aq, aqueduct; cc, central canal; ChAT, choline acetyltransferase; scp, superior cerebellar peduncle; TH, tyrosine hydroxylase.
Supplementary Figure 8 PVHMC4R neurons lie within the ARCPOMC→PVH efferent field
Fluorescent immunohistological analysis of MC4R-t2a-Cre::R26-loxSTOPlox-L10-GFP (green) and endogenous alpha-melanocyte-stimulating hormone (red) expression in the PVH revealed these two fields to be overlapping. Abbreviations: 3v, third ventricle. Scale bar = 100 μm.
Supplementary Figure 9 PVHMC4R→vlPAG or PVHMC4R→NTS/DMV neurons do not influence feeding behavior
a-b, in vivo optogenetic stimulation of PVHMC4R→vlPAG (a; n=4, Repeated measures ANOVA, main effect of interaction and treatment not significant, main effect of time (F(3,12)=76.00, p<0.0001) and PVHMC4R→NTS/DMV (b; n=5, Repeated measures ANOVA, main effect of interaction and treatment not significant, main effect of time (F(3,16)=20.61, p<0.0001) terminals does not promote satiety during dark-cycle feeding. c-e, PVHMC4R→LPBN (c), PVHMC4R→vlPAG (d), PVHMC4R→NTS/DMV (e) mice used for optogenetic feeding and RTPP studies were validated for fiber placement using histological sections. The approximate positions of the fibers is denoted by an X.
Supplementary Figure 10 PVHMC4R→LPBN photostimulation in the absence of ChR2-mCherry does not influence food intake
a, PVHMC4R neurons were transduced with cre-dependent GFP and optic fibers placed bilaterally over the LPBN. b, Photostimulation of PVHMC4R::GFP LPBN terminals had no effect on food intake during the dark-cycle, compared to the same mice without photostimulation (n=10, Repeated measures ANOVA, main effect of treatment and interaction not significant, main effect of time (F(3,36)=95.38, p<0.0001). c, following an overnight fast, as compared to same mice without photostimulation (n=10, Repeated measures ANOVA, main effect of treatment and interaction not significant, main effect of time (n=6, F(3,20)=132.6, p<0.0001). d, PVHMC4R neurons do not make synaptic contact with LPBNMC4R neurons. Channelrhodopsin-assisted circuit mapping between pre-synaptic PVHMC4R neurons (red) and putative post-synaptic LPBNMC4R neurons (green) in an Mc4r-t2a-Cre::R26-loxSTOPlox-L10-GFP mouse line revealed the absence of light-evoked EPSCs in all cells tested.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 (PDF 2818 kb)
Rights and permissions
About this article
Cite this article
Garfield, A., Li, C., Madara, J. et al. A neural basis for melanocortin-4 receptor–regulated appetite. Nat Neurosci 18, 863–871 (2015). https://doi.org/10.1038/nn.4011
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4011
This article is cited by
-
Advanced neurobiological tools to interrogate metabolism
Nature Reviews Endocrinology (2023)
-
Increased body weight in mice with fragile X messenger ribonucleoprotein 1 (Fmr1) gene mutation is associated with hypothalamic dysfunction
Scientific Reports (2023)
-
Adrenergic modulation of melanocortin pathway by hunger signals
Nature Communications (2023)
-
A sex-specific thermogenic neurocircuit induced by predator smell recruiting cholecystokinin neurons in the dorsomedial hypothalamus
Nature Communications (2023)
-
Endocannabinoids at the synapse and beyond: implications for neuropsychiatric disease pathophysiology and treatment
Neuropsychopharmacology (2023)