Abstract
We analyzed the progeny of individual neural stem cells (NSCs) of the mouse adult subependymal zone (SEZ) in vivo and found a markedly fast lineage amplification, as well as limited NSC self-renewal and exhaustion in a few weeks. We further unraveled the mechanisms of neuronal subtype generation, finding that a higher proportion of NSCs were dedicated to generate deep granule cells in the olfactory bulb and that larger clones were produced by these NSCs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Shook, B.A., Manz, D.H., Peters, J.J., Kang, S. & Conover, J.C. J. Neurosci. 32, 6947–6956 (2012).
Ninkovic, J., Mori, T. & Gotz, M. J. Neurosci. 27, 10906–10911 (2007).
Imayoshi, I. et al. Nat. Neurosci. 11, 1153–1161 (2008).
Codega, P. et al. Neuron 82, 545–559 (2014).
Bonaguidi, M.A. et al. Cell 145, 1142–1155 (2011).
Encinas, J.M. et al. Cell Stem Cell 8, 566–579 (2011).
Beckervordersandforth, R. et al. Cell Stem Cell 7, 744–758 (2010).
Mirzadeh, Z., Merkle, F.T., Soriano-Navarro, M., Garcia-Verdugo, J.M. & Alvarez-Buylla, A. Title. Cell Stem Cell 3, 265–278 (2008).
Ponti, G. et al. Proc. Natl. Acad. Sci. USA 110, E1045–E1054 (2013).
Merkle, F.T. et al. Nat. Neurosci. 17, 207–214 (2014).
Merkle, F.T., Mirzadeh, Z. & Alvarez-Buylla, A. Science 317, 381–384 (2007).
Lledo, P.M., Merkle, F.T. & Alvarez-Buylla, A. Trends Neurosci. 31, 392–400 (2008).
Mori, T. et al. Glia 54, 21–34 (2006).
DeCarolis, N.A. et al. Hippocampus 23, 708–719 (2013).
Ritsma, L. et al. Nature 507, 362–365 (2014).
Snippert, H.J. et al. Cell 143, 134–144 (2010).
Costa, M.R. et al. Development 138, 1057–1068 (2011).
Merkle, F.T. & Eggan, K. Cell Stem Cell 12, 656–668 (2013).
Lugert, S. et al. Nat. Commun. 3, 670 (2012).
Acknowledgements
We foremost would like to thank H. Clevers and H. Snippert for the Confetti mouse line. In addition, we are thankful to T. Özturk for excellent technical help and L. Dimou, A. Saghatelyan, S. Stricker and P. Malatesta for critical reading of the manuscript. We also gratefully acknowledge funding to J.N. from the German Research foundation (DFG) by the SFB 870 and SPP 1392 and to M.G. from the German Research foundation (DFG) by the SFB 870, Gö 640/11-1, the Munich Cluster for Systems Neurology (EXC 1010 SyNergy), the Helmholtz Foundation by the ICEMED Alliance and the European Research Council by an ERC Advanced Grant.
Author information
Authors and Affiliations
Contributions
F.C., J.M., J.N. and M.G. designed the experiments. F.C. and J.M. performed the experiments and analyzed data. E.V.B. provided technical assistance. F.T. performed mathematical modeling. F.C., J.N. and M.G. wrote the manuscript, to which F.T. contributed the mathematical modeling method section. All of the authors critically read and contributed comments to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
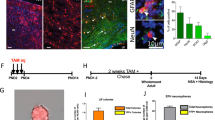
Supplementary Figure 1 Recombination within the neurogenic lineage is clonal and specific to NSCs.
(a) Relative abundance of distinct cell types within the recombined population, assessed 1.3 days (i.e. 32 hrs) after a single i.p. injection of 50μg/g of Tamoxifen (n=38 cells analyzed, from n=3 animals). (b) Example of initially recombined GFAP+ NSC with radial morphology (arrowheads point to radial GFAP+ process). (c) Representative TAP cluster positive for Ki67 (single focal section and orthogonal views in c´, c´´), lacking expression of Dcx and GFAP. (d) Graphs depicting the probability of observing activated clones as observed experimentally (blue; n=24 clones from n=54 hemispheres) and as predicted by mathematical modelling using the 10 μg/g Tm dose data set. (e) Predicted and the observed number of recombined NSCs (radial astrocytes; n=17,22,7,2 hemispheres analyzed per time point) at different time points, used to calculate the probabilities of clonal superimposition at different time points after induction. (f) Normalized observed (single data points) and simulated (curve) abundance of quiescent NSCs (qNSCs) over time; inset shows the deviation between predicted and observed values as a function of qNSC activation probability. Experimental data in d,f (inset) are shown as mean ± s.e.m. Simulation in f run using pc=0.0015 (black line) ± 0.0004 (gray shading) day−1. Scale bars 20μm.
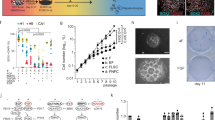
Supplementary Figure 2 Recombination within the neurogenic lineage is specific to NSCs and allows dissection of clonal expansion dynamics.
(a) Micrographs depicting a TAP doublet 3dpi. (b) Clone size and composition for all observed clones at the indicated time points (n=51 clones, from n=105 hemispheres. (c) Average (histograms) and individual (circles) clone size values per clone class, for each time point analyzed(n=2,7,1,2,12,9,3,2,3,7,3 clones, respectively). (d) Average number of regional bins (as defined in Fig. 1i) occupied by TAPs/NBs belonging to “emerging” and “mature” clones, across all time points (n=23,13 clones analyzed). (e) Two examples of NB (clone#1) and NB/TAP (clone#2) cluster distributions within the SEZ; sagittal views of sections corresponding to the indicated mediolateral positions (SEZ in yellow). (f) Average SEZ occupancy (% of SEZ-containing sagittal sections) by clusters of TAPs and NBs, for all clones (n=11,14 clones analyzed, respectively). (g) Subclonal birthdating paradigm (above) and number, spatial distribution and identity of birth-dated cells in a single clone (histogram). (h) Examples of clonally related cells belonging to young (left) and old NSC progeny cohorts(right). (i) Example of clustered BrdU/Ki67-labeled NSC and associated progeny. (j) Size of TAP-only, TAP/NB mixed and NB-only clusters/clones found in the indicated distinct regions. Y-axis values are the base-2 logarithm of individual cluster sizes (circles) and of the average cluster size (bars; n=5,5,10,15 clusters analyzed, respectively). Scale bars 20μm. Data in c, g are shown as mean ± s.e.m.
Supplementary Figure 3 Multiple cellular mechanisms underlay generation of neuronal diversity.
(a) Histogram showing the average total clonal size (red bars) and neuron number (blue bars) for clone types as defined by their neuronal composition (n=2,2,1,15,1,1,1,3,2 clones analyzed, respectively). (b) Metrics of OB subtype abundance, indicating the relative abundance of a given neuronal subtype in clones containing it (blue bars) and the abundance of the clones containing the various subtypes (green bars; n=22, 1, 7, 5, 5 clones analyzed, respectively). (c) Average size (red bars) and neuron number (blue bars) for clones containing one, two or three distinct neuronal subtypes (n=22, 5, 1 clones analyzed, respectively). Data are shown as mean ± s.e.m.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Table 1 (PDF 8398 kb)
Supplementary Table 2
Summary of all observed clones at different time points after the TM induction. (XLSX 42 kb)
Rights and permissions
About this article
Cite this article
Calzolari, F., Michel, J., Baumgart, E. et al. Fast clonal expansion and limited neural stem cell self-renewal in the adult subependymal zone. Nat Neurosci 18, 490–492 (2015). https://doi.org/10.1038/nn.3963
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.3963
This article is cited by
-
Dbx2, an Aging-Related Homeobox Gene, Inhibits the Proliferation of Adult Neural Progenitors
Stem Cell Reviews and Reports (2023)
-
RNA supply drives physiological granule assembly in neurons
Nature Communications (2022)
-
Multicolor strategies for investigating clonal expansion and tissue plasticity
Cellular and Molecular Life Sciences (2022)
-
Deciphering neural heterogeneity through cell lineage tracing
Cellular and Molecular Life Sciences (2021)
-
Momordica charantia polysaccharides modulate the differentiation of neural stem cells via SIRT1/Β-catenin axis in cerebral ischemia/reperfusion
Stem Cell Research & Therapy (2020)