Abstract
To examine the interaction between molecular, electrical and behavioral circadian rhythms, we combined optogenetic manipulation of suprachiasmatic nucleus (SCN) firing rate with bioluminescence imaging and locomotor activity monitoring. Manipulating firing rate reset circadian rhythms both ex vivo and in vivo, and this resetting required spikes and network communication. This suggests that SCN firing rate is fundamental to circadian pacemaking as both an input to and output of the molecular clockworks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Colwell, C.S. Nat. Rev. Neurosci. 12, 553–569 (2011).
Kuhlman, S.J. & McMahon, D.G. J. Biol. Rhythms 21, 470–481 (2006).
Brancaccio, M., Maywood, E.S., Chesham, J.E., Loudon, A.S. & Hastings, M.H. Neuron 78, 714–728 (2013).
Kudo, T. et al. J. Neurophysiol. 110, 1097–1106 (2013).
Freeman, G.M., Krock, R.M., Aton, S.J., Thaben, P. & Herzog, E.D. Neuron 78, 799–806 (2013).
Welsh, D.K., Takahashi, J.S. & Kay, S.A. Annu. Rev. Physiol. 72, 551–577 (2010).
Herzog, E.D., Takahashi, J.S. & Block, G.D. Nat. Neurosci. 1, 708–713 (1998).
Kuhlman, S.J., Silver, R., Le Sauter, J., Bult-Ito, A. & McMahon, D.G. J. Neurosci. 23, 1441–1450 (2003).
Nitabach, M.N., Blau, J. & Holmes, T.C. Cell 109, 485–495 (2002).
Yamaguchi, S. et al. Science 302, 1408–1412 (2003).
Welsh, D.K., Logothetis, D.E., Meister, M. & Reppert, S.M. Neuron 14, 697–706 (1995).
Schwartz, W.J., Gross, R.A. & Morton, M.T. Proc. Natl. Acad. Sci. USA 84, 1694–1698 (1987).
Goold, C.P. & Nicoll, R.A. Neuron 68, 512–528 (2010).
Sakai, K. Neuroscience 260, 249–264 (2014).
Ding, J.M. et al. Science 266, 1713–1717 (1994).
Daan, S. & Pittendrigh, C.S. J. Comp. Physiol. 106, 253–266 (1976).
Quintero, J.E. & McMahon, D.G. J. Neurophysiol. 82, 533–539 (1999).
Morin, L.P. & Allen, C.N. Brain Res. Brain Res. Rev. 51, 1–60 (2006).
Yoo, S.H. et al. Proc. Natl. Acad. Sci. USA 101, 5339–5346 (2004).
Evans, J.A., Leise, T.L., Castanon-Cervantes, O. & Davidson, A.J. Neuron 80, 973–983 (2013).
Atkins, N. et al. PLoS ONE 5, e12612 (2010).
Yizhar, O., Fenno, L.E., Davidson, T.J., Mogri, M. & Deisseroth, K. Neuron 71, 9–34 (2011).
Aravanis, A.M. et al. J. Neural Eng. 4, S143–S156 (2007).
Gamble, K.L., Allen, G.C., Zhou, T. & McMahon, D.G. J. Neurosci. 27, 12078–12087 (2007).
Ruan, G.X., Allen, G.C., Yamazaki, S. & McMahon, D.G. PLoS Biol. 6, e249 (2008).
Ruan, G.X., Gamble, K.L., Risner, M.L., Young, L.A. & McMahon, D.G. PLoS ONE 7, e38985 (2012).
Acknowledgements
We thank H. Iwamoto for assistance with the whole-cell recording experiments, and T. Page, C. Johnson and L. Funkhouser-Jones for helpful discussion and manuscript comments. This work was supported by US National Institutes of Health grants R01 EY015815 (D.G.M.), T32 MH064931 (J.R.J.) and F31 NS082213 (J.R.J.); US National Science Foundation Graduate Research Fellowship 0909667 (M.C.T.); and a Vanderbilt University Discovery Grant (D.G.M.).
Author information
Authors and Affiliations
Contributions
J.R.J., M.C.T. and D.G.M. prepared the paper. J.R.J. analyzed data. J.R.J. performed all in vitro and ex vivo experiments. J.R.J. and M.C.T. performed all in vivo experiments. J.R.J., M.C.T. and D.G.M. designed the experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
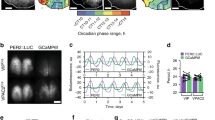
Supplementary Figure 1 Drd1a-Cre–driven ChR2 expression in the SCN.
(a) Low-magnification image of a representative Drd1a-ChR2 coronal brain section (n = 2) showing endogenous tdTomato fluorescence (magenta). Note that Drd1a-ChR2 is expressed strongly in the SCN; it is also expressed in the striatum, but shows minimal expression in the rest of the section (notably absent along the midline); scale bar, 1 mm. (b) Picture of a Drd1a-Cre brain section adapted from GENSAT’s Cre database at http://www.gensat.org/cre.jsp; note that Drd1a-Cre (brown) shows a comparable expression pattern to our low-magnification representative Drd1a-ChR2 brain section shown in (a); scale bar, 1 mm. (c) Representative 200 µm rostral-caudal serial coronal sections of the SCN and hypothalamus (n = 2) from a Drd1a-ChR2 mouse showing endogenous tdTomato fluorescence (magenta); scale bar 300 µm.
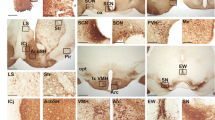
Supplementary Figure 2 Drd1a-driven optogenetic constructs are expressed in the SCN and can stimulate or inhibit SCN neurons.
(a) Immunohistochemical labeling of the SCN for AVP (cyan) and Drd1a-ChR2 (magenta); upper scale bar, 50 µm; lower scale bar, 5 µm; n = 5 slices. (b) Immunohistochemical labeling of the SCN for VIP (cyan) and Drd1a-ChR2 (magenta); upper scale bar, 50 µm; lower scale bar, 5 µm; n = 5 slices. ChR2 is shown to be expressed across the SCN, with 93.71 ± 1.41% of AVP-positive neurons and 89.65 ± 2.31% of VIP-positive neurons colocalized with ChR2. (c) Action potentials are generated by blue light stimulation in 92.68 ± 2.00% of Drd1a-ChR2 SCN neurons; n = 6 slices, 53 cells (top) and inhibited by yellow light in 84.21 ± 3.84% of Drd1a-NpHR SCN neurons; n = 5 slices, 37 cells (bottom). (d) The percentage of stimulus pulses that generated action potentials in Drd1a-ChR2 SCN neurons in response to a train of blue light pulses is near-100% up to approximately 12 Hz; at higher stimulation frequencies, spike fidelity declines; n = 4 cells per frequency. (e) Representative firing rate histogram showing that Drd1a-ChR2 neurons can be driven at 10 Hz for over an hour; after the conclusion of stimulation, firing rate returns to baseline; n = 3 cells. (f) Representative firing rate histogram showing that Drd1a-NpHR neurons can be inhibited for over an hour; after the conclusion of inhibition, firing rate returns to baseline; n = 3 cells.
Supplementary Figure 3 Optogenetic stimulation of organotypic Drd1a-ChR2 SCN slices results in widespread c-Fos activation.
(a) (Left) Representative immunohistochemical labeling of ChR2+ SCN slices for cFos (cyan) and Drd1a-ChR2 (magenta); (Right) Representative immunohistochemical labeling of ChR2- (control) SCN slices for cFos (cyan); 3V = third ventricle; OC = optic chiasm; dashed white line = approximate outline of SCN as determined by Drd1a-ChR2 fluorescence (ChR2+) or cell density (ChR2-); scale bar, 50 µm; n = 3 slices per genotype. (b) Quantification of the number of cFos-positive cells in the SCN of ChR2+ or ChR2- SCN slices; Student’s t-test, p < 0.001. (c) Quantification of the number of ChR2-positive cells that are also cFos-positive (87.83 ± 0.97%) and the number of cFos-positive cells that are also ChR2-positive (92.17 ± 2.34%) after optogenetic stimulation. Data are presented as means ± SEM.
Supplementary Figure 4 Optogenetic manipulation of the SCN ex vivo produces changes in period that are correlated with changes in phase.
(a) Period response curve depicting changes of period in response to ChR2-mediated stimulation in the same slices depicted in Figs. 1a and 1b; dashed line, Gaussian fit, r2 = 0.7466. (b) Changes in phase and changes in period in response to ChR2-mediated stimulation are positively correlated; Pearson’s r, r2 = 0.9342, p<0.001. For (a-b), blue dots, ChR2+, n = 23 slices, 20 animals; black dots, ChR2-, n = 11 slices, 11 animals. (c) Period response curve depicting changes of period in response to NpHR-mediated inhibition in the same slices depicted in Figs. 1c and 1d; dashed line, Gaussian fit, r2 = 0.6302. (d) Changes in phase and changes in period in response to NpHR-mediated inhibition are positively correlated; Pearson’s r, r2 = 0.8565, p<0.001. For (c-d), yellow dots, NpHR+, n = 19 slices, 14 animals; black dots, NpHR-, n = 11 slices, 9 animals.
Supplementary Figure 5 Pharmacological blockade of action potential generation or VIP signaling ablates period changes induced by ChR2 stimulation.
Changes in period of PER2∷LUC rhythms from slices in Fig. 2 in response to combinations of ChR2 stimulation (470 nm, 1 hour, 8 Hz; ‘STIM’), TTX, and/or VIPX; One-Way ANOVA with post-hoc HSD, F: 7.92, p = 0.0004. Data are presented as means ± SEM.
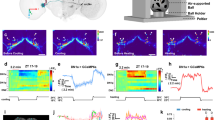
Supplementary Figure 6 In vivo optogenetic stimulation localized to the SCN.
(a) Schematic representing the brain area illuminated by 470 nm light of sufficient intensity to excite ChR2 as delivered through the stereotaxically implanted fiber optic used in our experiments (400 µM diameter, 0.39 NA). Measured light intensity at the fiber tip of 10.8 mW. The cone of blue shading depicts the distance from the fiber tip at which the irradiance is ≥ 1 mW/mm2, which is the estimated ChR2 activation threshold (see (b)); scale bar, 1 mm. (b) Calculated linear (top) and log (bottom) irradiance values attenuate rapidly as distance from fiber tip increases. Calculations were based on a measured light intensity at the fiber tip of 10.8 mW and were performed using the “Brain tissue light transmission calculator” available at www.optogenetics.org. Dashed line in the bottom graph represents an irradiance of 1 mW/mm2. (c) Representative brain slices (n = 3) from control (top) and ChR2+ (bottom) animals implanted with a fiber optic cannula dipped in blue dye and immediately perfused and sectioned; asterisk depicts approximate location of fiber tip; scale bar, 1 mm.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 1775 kb)
Rights and permissions
About this article
Cite this article
Jones, J., Tackenberg, M. & McMahon, D. Manipulating circadian clock neuron firing rate resets molecular circadian rhythms and behavior. Nat Neurosci 18, 373–375 (2015). https://doi.org/10.1038/nn.3937
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.3937
This article is cited by
-
In vivo recording of the circadian calcium rhythm in Prokineticin 2 neurons of the suprachiasmatic nucleus
Scientific Reports (2023)
-
Human blood serum proteome changes after 6 hours of sleep deprivation at night
Sleep Science and Practice (2021)
-
Circadian neurons in the paraventricular nucleus entrain and sustain daily rhythms in glucocorticoids
Nature Communications (2021)
-
Data-independent acquisition method for ubiquitinome analysis reveals regulation of circadian biology
Nature Communications (2021)
-
Circadian Regulation of GluA2 mRNA Processing in the Rat Suprachiasmatic Nucleus and Other Brain Structures
Molecular Neurobiology (2021)