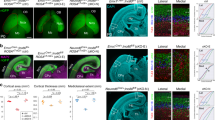
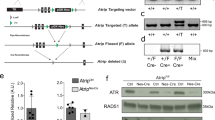
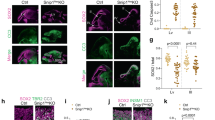
Abstract
The rapid proliferation of progenitors during neurogenesis requires a stringent genomic maintenance program to ensure transmission of genetic fidelity. However the essential factors that govern neural progenitor genome integrity are unknown. Here we report that conditional inactivation of mouse TopBP1, a protein linked to DNA replication, and a key activator of the DNA damage response kinase ATR (ataxia telangiectasia and rad3-related) is critical for maintenance of early-born neural progenitors. During cortical development TopBP1 prevented replication-associated DNA damage in Emx1-progenitors which otherwise resulted in profound tissue ablation. Notably, disrupted neurogenesis in TopBP1-depleted tissues was substantially rescued by inactivation of p53 but not of ATM. Our data establish that TopBP1 is essential for preventing replication-associated DNA strand breaks, but is not essential per se for DNA replication. Thus, TopBP1 is crucial for maintaining genome integrity in the early progenitors that drive neurogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Branzei, D. & Foiani, M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11, 208–219 (2010).
Jackson, S.P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
McKinnon, P.J. & Caldecott, K.W. DNA strand break repair and human genetic disease. Annu. Rev. Genomics Hum. Genet. 8, 37–55 (2007).
Tercero, J.A., Longhese, M.P. & Diffley, J.F. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11, 1323–1336 (2003).
McKinnon, P.J. DNA repair deficiency and neurological disease. Nat. Rev. Neurosci. 10, 100–112 (2009).
Balestrini, A., Cosentino, C., Errico, A., Garner, E. & Costanzo, V. GEMC1 is a TopBP1-interacting protein required for chromosomal DNA replication. Nat. Cell Biol. 12, 484–491 (2010).
Garcia, V., Furuya, K. & Carr, A.M. Identification and functional analysis of TopBP1 and its homologs. DNA Repair (Amst.) 4, 1227–1239 (2005).
Kumagai, A., Lee, J., Yoo, H.Y. & Dunphy, W.G. TopBP1 activates the ATR-ATRIP complex. Cell 124, 943–955 (2006).
Kumagai, A., Shevchenko, A. & Dunphy, W.G. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 140, 349–359 (2010).
Sansam, C.L. et al. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev. 24, 183–194 (2010).
Mäkiniemi, M. et al. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J. Biol. Chem. 276, 30399–30406 (2001).
Yamane, K., Kawabata, M. & Tsuruo, T. A DNA-topoisomerase-II-binding protein with eight repeating regions similar to DNA-repair enzymes and to a cell-cycle regulator. Eur. J. Biochem. 250, 794–799 (1997).
Yamane, K., Wu, X. & Chen, J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol. Cell. Biol. 22, 555–566 (2002).
Yu, X., Chini, C.C., He, M., Mer, G. & Chen, J. The BRCT domain is a phospho-protein binding domain. Science 302, 639–642 (2003).
Zegerman, P. & Diffley, J.F. DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst.) 8, 1077–1088 (2009).
Remus, D. & Diffley, J.F. Eukaryotic DNA replication control: lock and load, then fire. Curr. Opin. Cell Biol. 21, 771–777 (2009).
Cimprich, K.A. & Cortez, D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9, 616–627 (2008).
Liu, S. et al. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol. Cell. Biol. 26, 6056–6064 (2006).
Mordes, D.A., Glick, G.G., Zhao, R. & Cortez, D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 22, 1478–1489 (2008).
Navadgi-Patil, V.M. & Burgers, P.M. A tale of two tails: activation of DNA damage checkpoint kinase Mec1/ATR by the 9–1-1 clamp and by Dpb11/TopBP1. DNA Repair (Amst.) 8, 996–1003 (2009).
Jeon, Y. et al. TopBP1 deficiency causes an early embryonic lethality and induces cellular senescence in primary cells. J. Biol. Chem. 286, 5414–5422 (2011).
Liu, Q. et al. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 14, 1448–1459 (2000).
Brown, E.J. & Baltimore, D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 14, 397–402 (2000).
Delacroix, S., Wagner, J.M., Kobayashi, M., Yamamoto, K. & Karnitz, L.M. The Rad9-Hus1-Rad1 (9–1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 21, 1472–1477 (2007).
Furuya, K., Poitelea, M., Guo, L., Caspari, T. & Carr, A.M. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev. 18, 1154–1164 (2004).
Lee, J. & Dunphy, W.G. Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks. Mol. Biol. Cell 21, 926–935 (2010).
Cortez, D., Guntuku, S., Qin, J. & Elledge, S.J. ATR and ATRIP: partners in checkpoint signaling. Science 294, 1713–1716 (2001).
Greer, D.A., Besley, B.D., Kennedy, K.B. & Davey, S. hRad9 rapidly binds DNA containing double-strand breaks and is required for damage-dependent topoisomerase II beta binding protein 1 focus formation. Cancer Res. 63, 4829–4835 (2003).
Cotta-Ramusino, C. et al. A DNA damage response screen identifies RHINO, a 9–1-1 and TopBP1 interacting protein required for ATR signaling. Science 332, 1313–1317 (2011).
Gong, Z., Kim, J.E., Leung, C.C., Glover, J.N. & Chen, J. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol. Cell 37, 438–446 (2010).
Leung, C.C., Gong, Z., Chen, J. & Glover, J.N. Molecular basis of BACH1/FANCJ recognition by TopBP1 in DNA replication checkpoint control. J. Biol. Chem. 286, 4292–4301 (2011).
Murga, M. et al. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat. Genet. 41, 891–898 (2009).
Ruzankina, Y. et al. Tissue regenerative delays and synthetic lethality in adult mice after combined deletion of Atr and Trp53. Nat. Genet. 41, 1144–1149 (2009).
Götz, M. & Huttner, W.B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777–788 (2005).
Molyneaux, B.J., Arlotta, P., Menezes, J.R. & Macklis, J.D. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 8, 427–437 (2007).
Caviness, V.S. Jr., Nowakowski, R.S. & Bhide, P.G. Neocortical neurogenesis: morphogenetic gradients and beyond. Trends Neurosci. 32, 443–450 (2009).
Gorski, J.A. et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 22, 6309–6314 (2002).
Chou, S.J., Perez-Garcia, C.G., Kroll, T.T. & O'Leary, D.D. Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat. Neurosci. 12, 1381–1389 (2009).
Lee, Y. et al. The genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Nat. Neurosci. 12, 973–980 (2009).
Shull, E.R. et al. Differential DNA damage signaling accounts for distinct neural apoptotic responses in ATLD and NBS. Genes Dev. 23, 171–180 (2009).
Lee, Y., Chong, M.J. & McKinnon, P.J. Ataxia telangiectasia mutated-dependent apoptosis after genotoxic stress in the developing nervous system is determined by cellular differentiation status. J. Neurosci. 21, 6687–6693 (2001).
Rogakou, E.P., Boon, C., Redon, C. & Bonner, W.M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146, 905–916 (1999).
Katyal, S. et al. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 26, 4720–4731 (2007).
Arai, Y. et al. Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nat. Commun. 2, 154 (2011).
Dehay, C. & Kennedy, H. Cell-cycle control and cortical development. Nat. Rev. Neurosci. 8, 438–450 (2007).
Pulvers, J.N. & Huttner, W.B. Brca1 is required for embryonic development of the mouse cerebral cortex to normal size by preventing apoptosis of early neural progenitors. Development 136, 1859–1868 (2009).
Lee, Y. et al. ATR maintains select progenitors during nervous system development. EMBO J. 31, 1177–1189 (2012).
Herzog, K.H., Chong, M.J., Kapsetaki, M., Morgan, J.I. & McKinnon, P.J. Requirement for ATM in ionizing radiation-induced cell death in the developing central nervous system. Science 280, 1089–1091 (1998).
Lee, Y., Barnes, D.E., Lindahl, T. & McKinnon, P.J. Defective neurogenesis resulting from DNA ligase IV deficiency requires ATM. Genes Dev. 14, 2576–2580 (2000).
Orii, K.E., Lee, Y., Kondo, N. & McKinnon, P.J. Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc. Natl. Acad. Sci. USA 103, 10017–10022 (2006).
Acknowledgements
We thank the Hartwell Center for biotech support, the Transgenic Core Facility for blastocyst injections and chimera production and the ARC for animal husbandry. P.J.M. is supported by the US National Institutes of Health (NS-37956, CA-21765), a Cancer Center support grant (P30 CA21765) and the American Lebanese and Syrian Associated Charities of St. Jude Children's Research Hospital. S.K. is a Neoma Boadway Academic Programs Endowed Fellow.
Author information
Authors and Affiliations
Contributions
Y.L., S.K. and H.R.R. performed all experiments characterizing the Topbp1 mutant mouse and contributed to writing the manuscript. S.M.D. contributed the analysis of the Lig4 and Xrcc1 conditional mutant mice. J.Z. generated western blot data. P.J.M. was project leader and produced the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 (PDF 10508 kb)
Rights and permissions
About this article
Cite this article
Lee, Y., Katyal, S., Downing, S. et al. Neurogenesis requires TopBP1 to prevent catastrophic replicative DNA damage in early progenitors. Nat Neurosci 15, 819–826 (2012). https://doi.org/10.1038/nn.3097
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.3097
This article is cited by
-
SNIP1 and PRC2 coordinate cell fates of neural progenitors during brain development
Nature Communications (2023)
-
Microcephalin 1/BRIT1-TRF2 interaction promotes telomere replication and repair, linking telomere dysfunction to primary microcephaly
Nature Communications (2020)
-
Sonic hedgehog accelerates DNA replication to cause replication stress promoting cancer initiation in medulloblastoma
Nature Cancer (2020)
-
Yin Yang 1 sustains biosynthetic demands during brain development in a stage-specific manner
Nature Communications (2019)
-
Neural blastocyst complementation enables mouse forebrain organogenesis
Nature (2018)