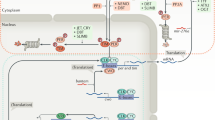
Abstract
Circadian clocks control a variety of neuronal, behavioral and physiological responses, via transcriptional regulation of an appreciable portion of the genome. We describe the complex communication network between the brain-specific central clock and the tissue-specific peripheral clocks that serve to synchronize the organism to both external and internal demands. In addition, we discuss and speculate on how epigenetic processes are involved in creating transcriptional environments that are permissive to tissue-specific gene expression programs, which work in concert with the circadian machinery. Accumulating data show that chromatin remodeling events may be critical for providing specificity and plasticity in circadian regulation, and metabolic cues may be involved in directing such epigenetic events. A detailed understanding of the communication cues between the central and peripheral clocks is crucial for a more complete understanding of the circadian system and the several levels of control that are implicated in maintaining biological timekeeping.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Reppert, S.M. & Weaver, D.R. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 63, 647–676 (2001).
Welsh, D.K., Takahashi, J.S. & Kay, S.A. Suprachiasmatic nucleus: cell autonomy and network properties. Annu. Rev. Physiol. 72, 551–577 (2010).
Bechtold, D.A., Gibbs, J.E. & Loudon, A.S. Circadian dysfunction in disease. Trends Pharmacol. Sci. 31, 191–198 (2010).
Sahar, S. & Sassone-Corsi, P. Metabolism and cancer: the circadian clock connection. Nat. Rev. Cancer 9, 886–896 (2009).
Thresher, R.J. et al. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science 282, 1490–1494 (1998).
Eckel-Mahan, K. & Sassone-Corsi, P. Metabolism control by the circadian clock and vice versa. Nat. Struct. Mol. Biol. 16, 462–467 (2009).
Gallego, M. & Virshup, D.M. Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 8, 139–148 (2007).
Aton, S.J., Colwell, C.S., Harmar, A.J., Waschek, J. & Herzog, E.D. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat. Neurosci. 8, 476–483 (2005).
Kalsbeek, A., Fliers, E., Hofman, M.A., Swaab, D.F. & Buijs, R.M. Vasopressin and the output of the hypothalamic biological clock. J. Neuroendocrinol. 22, 362–372 (2010).
Tousson, E. & Meissl, H. Suprachiasmatic nuclei grafts restore the circadian rhythm in the paraventricular nucleus of the hypothalamus. J. Neurosci. 24, 2983–2988 (2004).
Ralph, M.R., Foster, R.G., Davis, F.C. & Menaker, M. Transplanted suprachiasmatic nucleus determines circadian period. Science 247, 975–978 (1990).
Akhtar, R.A. et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 12, 540–550 (2002).
Cheng, M.Y. et al. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature 417, 405–410 (2002).
Kramer, A. et al. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science 294, 2511–2515 (2001).
Abe, K., Kroning, J., Greer, M.A. & Critchlow, V. Effects of destruction of the suprachiasmatic nuclei on the circadian rhythms in plasma corticosterone, body temperature, feeding and plasma thyrotropin. Neuroendocrinology 29, 119–131 (1979).
Balsalobre, A. et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289, 2344–2347 (2000).
Welsh, D.K., Logothetis, D.E., Meister, M. & Reppert, S.M. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14, 697–706 (1995).
Yamazaki, S. et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science 288, 682–685 (2000).
Panda, S. et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320 (2002).
Yan, J., Wang, H., Liu, Y. & Shao, C. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLOS Comput. Biol. 4, e1000193 (2008).
Baggs, J.E. et al. Network features of the mammalian circadian clock. PLoS Biol. 7, e52 (2009).
Ueda, H.R. et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 37, 187–192 (2005).
Travnickova-Bendova, Z., Cermakian, N., Reppert, S.M. & Sassone-Corsi, P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc. Natl. Acad. Sci. USA 99, 7728–7733 (2002).
Nader, N., Chrousos, G.P. & Kino, T. Circadian rhythm transcription factor CLOCK regulates the transcriptional activity of the glucocorticoid receptor by acetylating its hinge region lysine cluster: potential physiological implications. FASEB J. 23, 1572–1583 (2009).
Duffield, G.E. et al. A role for Id2 in regulating photic entrainment of the mammalian circadian system. Curr. Biol. 19, 297–304 (2009).
Kornmann, B., Schaad, O., Bujard, H., Takahashi, J.S. & Schibler, U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 5, e34 (2007).
Duffield, G.E. et al. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr. Biol. 12, 551–557 (2002).
Crosio, C., Cermakian, N., Allis, C.D. & Sassone-Corsi, P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat. Neurosci. 3, 1241–1247 (2000).
Etchegaray, J.P., Lee, C., Wade, P.A. & Reppert, S.M. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421, 177–182 (2003).
Nakahata, Y. et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340 (2008).
Cheung, P. et al. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5, 905–915 (2000).
Tu, H. et al. Dominant role of GABAB2 and Gβγ for GABAB receptor-mediated-ERK1/2/CREB pathway in cerebellar neurons. Cell. Signal. 19, 1996–2002 (2007).
Ding, J.M. et al. Resetting the biological clock: mediation of nocturnal circadian shifts by glutamate and NO. Science 266, 1713–1717 (1994).
Impey, S. et al. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron 34, 235–244 (2002).
Yujnovsky, I., Hirayama, J., Doi, M., Borrelli, E. & Sassone-Corsi, P. Signaling mediated by the dopamine D2 receptor potentiates circadian regulation by CLOCK:BMAL1. Proc. Natl. Acad. Sci. USA 103, 6386–6391 (2006).
Doi, M., Hirayama, J. & Sassone-Corsi, P. Circadian regulator CLOCK is a histone acetyltransferase. Cell 125, 497–508 (2006).
Ripperger, J.A. & Schibler, U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 38, 369–374 (2006).
Hirayama, J. et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 450, 1086–1090 (2007).
Asher, G. et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134, 317–328 (2008).
Alenghat, T. et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature 456, 997–1000 (2008).
Katada, S. & Sassone-Corsi, P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat. Struct. Mol. Biol. (in the press).
Etchegaray, J.P. et al. The polycomb group protein EZH2 is required for mammalian circadian clock function. J. Biol. Chem. 281, 21209–21215 (2006).
Roozendaal, B. et al. Membrane-associated glucocorticoid activity is necessary for modulation of long-term memory via chromatin modification. J. Neurosci. 30, 5037–5046 (2010).
Chwang, W.B., Arthur, J.S., Schumacher, A. & Sweatt, J.D. The nuclear kinase mitogen- and stress-activated protein kinase 1 regulates hippocampal chromatin remodeling in memory formation. J. Neurosci. 27, 12732–12742 (2007).
Gao, J. et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466, 1105–1109 (2010).
Nakahata, Y., Sahar, S., Astarita, G., Kaluzova, M. & Sassone-Corsi, P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654–657 (2009).
Ramsey, K.M. et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324, 651–654 (2009).
Lamia, K.A. et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326, 437–440 (2009).
Womac, A.D., Burkeen, J.F., Neuendorff, N., Earnest, D.J. & Zoran, M.J. Circadian rhythms of extracellular ATP accumulation in suprachiasmatic nucleus cells and cultured astrocytes. Eur. J. Neurosci. 30, 869–876 (2009).
Belden, W.J., Loros, J.J. & Dunlap, J.C. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol. Cell 25, 587–600 (2007).
Acknowledgements
We thank all members of our laboratory for discussions and support. Work in the laboratory is supported by the US National Institute of Health and the INSERM (Institut National de la Santé et la Recherche Médicale), France.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Masri, S., Sassone-Corsi, P. Plasticity and specificity of the circadian epigenome. Nat Neurosci 13, 1324–1329 (2010). https://doi.org/10.1038/nn.2668
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.2668
This article is cited by
-
Deficiency of circadian clock gene Bmal1 exacerbates noncanonical inflammasome-mediated pyroptosis and lethality via Rev-erbα-C/EBPβ-SAA1 axis
Experimental & Molecular Medicine (2024)
-
Intertwining roles of circadian and metabolic regulation of the innate immune response
Seminars in Immunopathology (2022)
-
Sirtuins and the circadian clock interplay in cardioprotection: focus on sirtuin 1
Cellular and Molecular Life Sciences (2021)
-
Circadian Rhythms in Environmental Health Sciences
Current Environmental Health Reports (2020)
-
Disruption of the Molecular Circadian Clock and Cancer: An Epigenetic Link
Biochemical Genetics (2020)